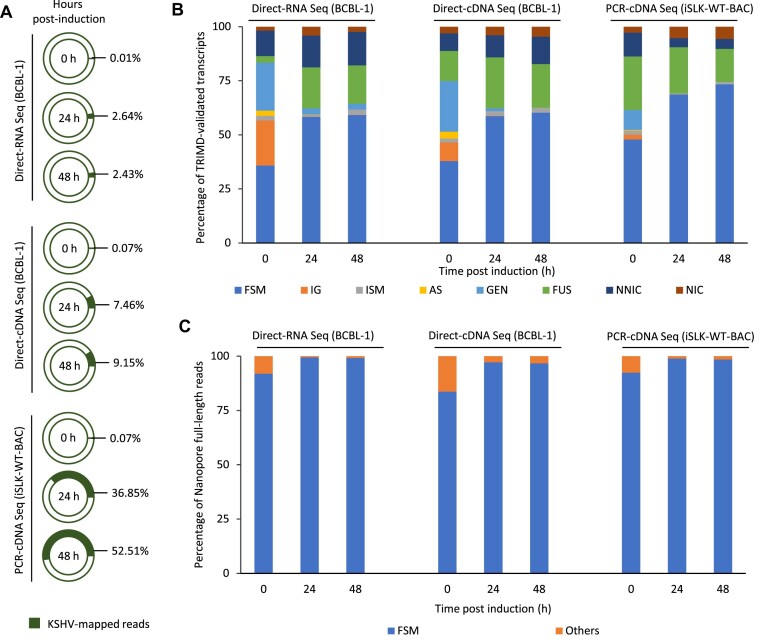
Figure 8.
Comparison of TRIMD-identified transcripts and ONT sequencing reads. (A) Percentage of total reads that aligned to KSHV as generated from the direct-RNA and direct-cDNA sequencing of BCBL-1 cells and PCR-cDNA sequencing of iSLK-WT-BAC16 cells, before and after induction. (B) TRIMD-identified transcripts classified into full splice match (FSM), incomplete splice match (ISM), intergenic (IG), antisense (AS), genic (GEN), fusion genes (FUS), novel in catalogue (NIC) and novel not in catalogue (NNIC) transcripts with reference to full-length KSHV reads obtained from different platforms of nanopore sequencing of BCBL-1 and iSLK-WT-BAC16 cells. (C) Full-length KSHV transcripts processed from nanopore sequencing platforms of BCBL-1 cells and iSLK-WT-BAC16 cells classified into full splice match (FSM) and Others (including ISM, GEN, FUS, NIC, NNIC and AS) with reference to TRIMD-identified lytic transcripts.