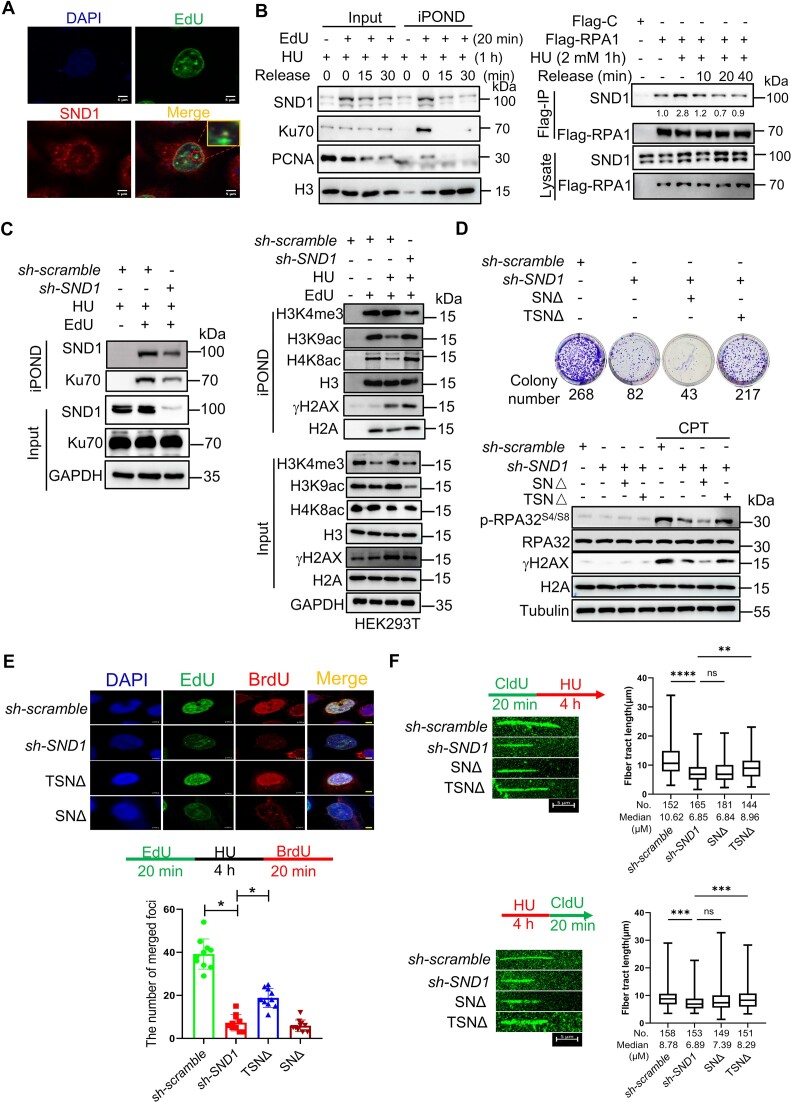
Figure 6.
SN domain mediated the regulation of stability and restart of replication forks by SND1. (A) The association of SND1 with nascent DNA in HeLa cells treated with CPT. EdU incorporation was performed as previously publications. Representative images with scale bars (5 μm) are shown. (B) Western blot of nascent DNA-associated proteins at stalled replication forks. Nascent DNA-associated proteins were isolated in HEK293T cells treated with HU for 1 h and followed by releasing into fresh media for indicated time using iPOND approach, and the resulted proteins were detected using specific indicated antibodies (left panel). Western blot of RPA1 immunoprecipitation in HEK293T cells treated with HU for 1 hour and followed by releasing into fresh media for indicated time. Copurified proteins were detected using specific antibodies. The number represented the quantification of SND1 protein against to Flag-RPA1 (right panel). (C) Western blot for H3K9ac, H4K8ac, γH2AX and Ku70 at the nascent DNA in SND1 deficient HEK293T cells. (D) Complement of SN domain of SND1partially rescue the viability of HeLa cells lacking endogenous SND1 protein in response to CPT treatment (Upper panel). Western blot for phospho-RPA32S4/S8 and γH2AX in HeLa cells in response to CPT treatment (Lower panel). (E) BrdU and EdU were incorporated into nascent DNA for observing the replication forks foci formation and restart of replication forks by immunofluorescence method. The histogram showed the number of replication foci formation in indicated HeLa cells. Scale bars, 5 μm. (F) DNA fiber assay for exploring the function of SND1 and its truncated protein on the progression (upper panel) and restart (lower panel) of DNA replication forks. The histogram showed the alteration in the length of nascent DNA in HeLa. All experiments were independently repeated three times, *P < 0.05.