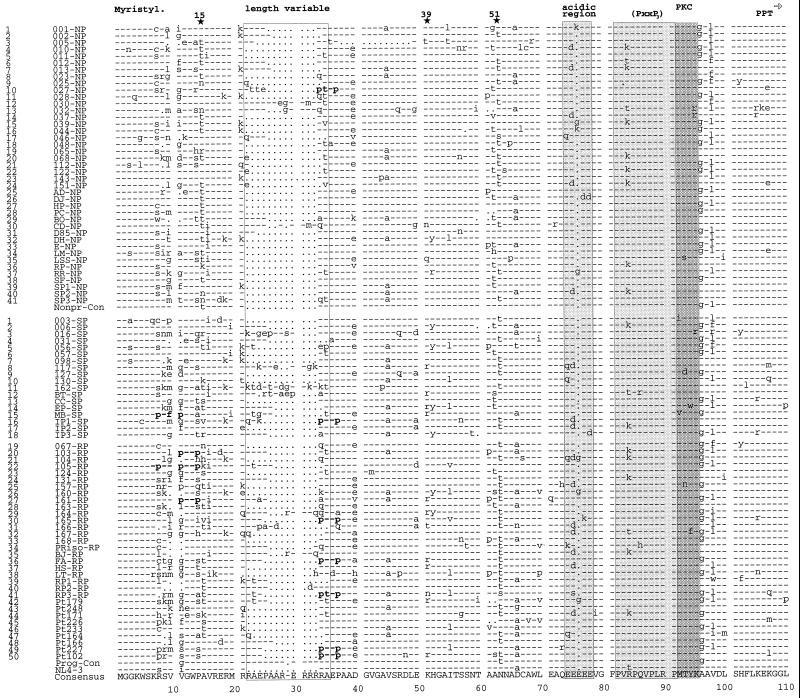
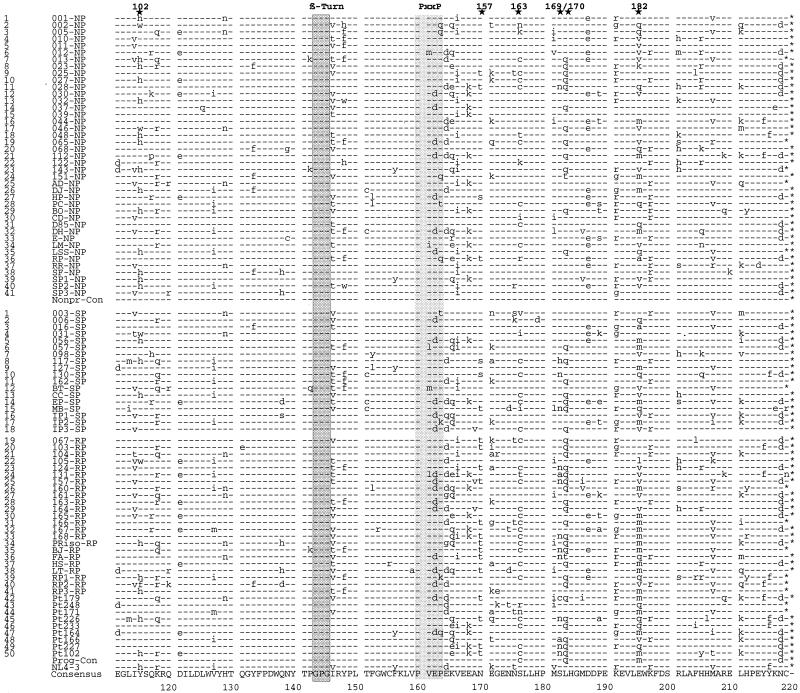
FIG. 1.
Alignment of Nef protein sequences derived from HIV-1-infected individuals with different rates of disease progression. Representative consensus Nef protein sequences from 41 NPs, 18 SPs, and 32 RPs were aligned. Nef protein sequences were derived from the present study (NPs 1 to 28, SPs 1 to 15, and RPs 19 to 38) and from studies by Huang et al. (15) (NPs 29 to 38), Michael et al. (27) (NPs 39 to 41, SPs 16 to 18, and RPs 39 to 41), and Shugars et al. (38) (patients 42 to 50). The consensus amino acid sequence is shown at the bottom. The NP and progressor consensus sequences and the HIV-1 NL4-3 sequence are also indicated. The first column indicates the index number for all NPs and progressors (SPs and RPs combined) in the study. In the second column, the first two to four numbers or letters specify the individual patient, and the last letter(s) specifies the progression grouping. Some conserved sequence elements are indicated schematically, and additional PxxP motifs close to the N terminus are in boldface. The position of the polypurine tract (PPT) and the start of the 3′ LTR (➩) are also indicated. Stars above the alignment indicate positions where amino acid variations between the different progressor groups are observed; the number gives the corresponding amino acid position in the NL4-3 Nef. Dashes indicate identity with the consensus sequence, dots indicate gaps introduced to optimize the alignment, and asterisks indicate stop codons.