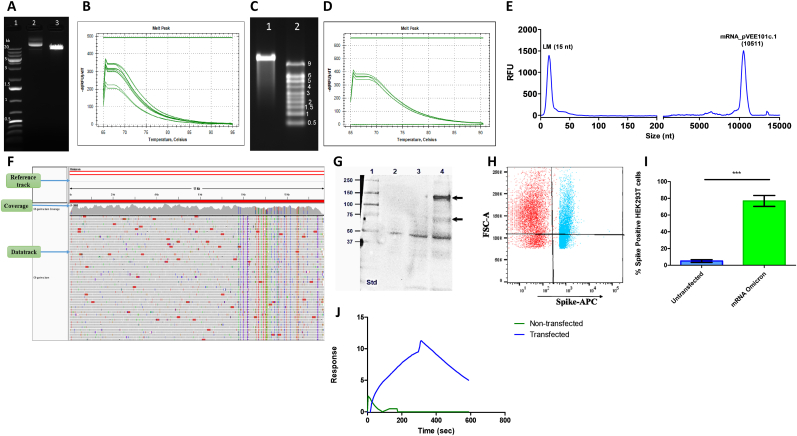
Fig. 4.
Plasmid DNA and mRNA characterization.
(A) Ethidium bromide-stained agarose gel image of plasmid DNA pVEE 101c.1, Lane:1 1 kb plus DNA ladder. Lane 2: pVEE 101c.1 uncut, Lane 3: pVEE 101c.1 linearized plasmid.
(B) Melt peaks of the pVEE 101c.1 plasmid DNA sample from qPCR reaction.
(C) Ethidium bromide stained MOPS gel image of mRNA. Lane 1: mRNA encoded by pVEE 101c.1, Lane 2: RNA Millennium 9 kb ladder.
(D) Melt peaks of mRNA sample on qPCR for the detection of pVEE101c.1 plasmid DNA contamination.
(E) Capillary electrophoresis profile of mRNA on 5200 fragment analyzer system.
(F) Snapshot of NGS data alignment of mRNA with the reference sequence file.
(G) The whole cell extract of mRNA transfected and non-transfected HEK293T was probed using anti-SARS-CoV-2 spike mouse monoclonal IgG and detected using HRP-conjugated goat anti-mouse IgG, lane 1: marker, lane 2. non-transfected, lane 3: mRNA from empty vector (without antigenic gene), and lane 4: mRNA encoding Omicron spike.
(H) Shows the scatter plot of FACS for non-transfected and mRNA-transfected HEK293T cells.
(I) The % of spike-positive HEK 293T cells is plotted as an average of three independent sets of experiments for both transfected and un-transfected cells. The error bar represents the standard deviation. ***p < 0.05
(J) The whole cell extract of transfected and non-transfected cells was analyzed on Biacore 8K + by immobilizing ACE-2 on the CM5 chip. Shown is one of the representative sensorgrams of the transfected and non-transfected cell lysate.