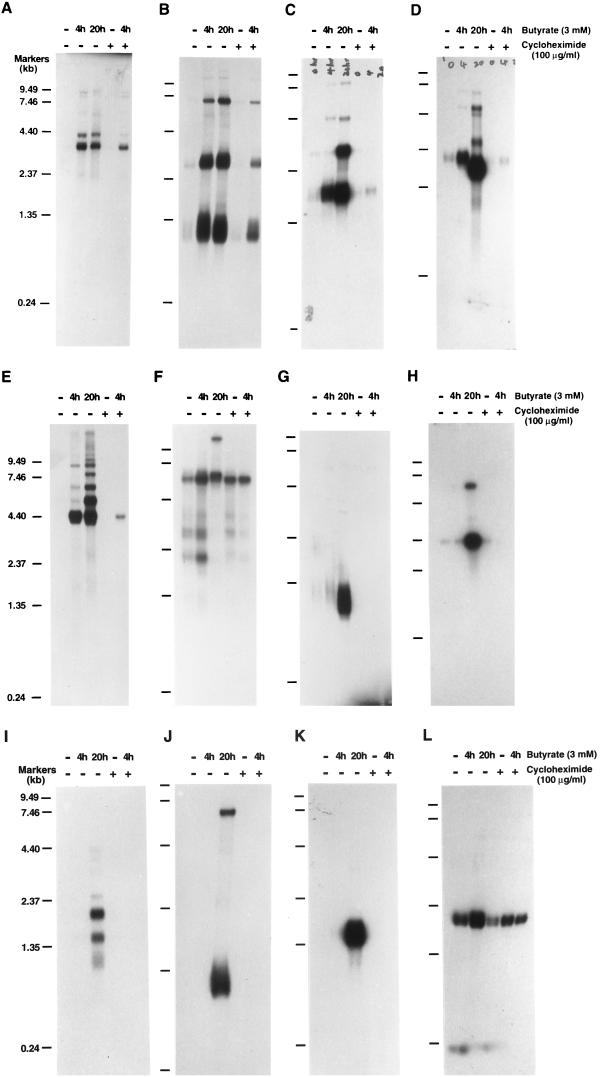
FIG. 3.
Northern analysis of KSHV IE mRNAs. Poly(A)+ RNA was isolated from nonstimulated BC-1 cells and BC-1 cells that had been treated with sodium butyrate for 4 or 20 h in the absence or presence of cycloheximide as indicated above each lane. RNA from BC-1 cells that were treated with cycloheximide for 8 h but not induced was also included. These RNA samples were separated on a 1.0% agarose-formaldehyde gel and transferred onto Nytran membranes. The membranes were probed with different 32P-labeled single-stranded DNA probes that were prepared by asymmetric PCR as described in Materials and Methods. The probes are as follows: single-stranded DNA complementary to nucleotides 73682 to 73956 in the KSHV genome within ORF50 (A); single-stranded DNA complementary to nucleotides 73956 to 73682, the antisense sequence of ORF50 (B); single-stranded DNA complementary to nucleotides 68502 to 68228 within ORF45 (C); single-stranded DNA complementary to nucleotides 22751 to 23073 within ORF K4.2 (D); single-stranded DNA complementary to nucleotides 50016 to 50261 (E); random priming-labeled ORF73 (LANA) cDNA (nucleotides 123809 to 127297) (F); random priming-labeled ORF K2 (vIL-6) cDNA (nucleotides 17261 to 17875) (G); random priming-labeled ORF59 cDNA (nucleotides 95549 to 96739) (H); random priming-labeled K3 cDNA (nucleotides 18608 to 19609) (I); random priming-labeled K5 cDNA (nucleotides 25713 to 26483) (J); random priming-labeled ORF57 cDNA (nucleotides 82717 to 83544) (K); and β-actin cDNA as a control for RNA integrity and loading (L). Molecular markers were an 0.24- to 9.5-kb RNA ladder.