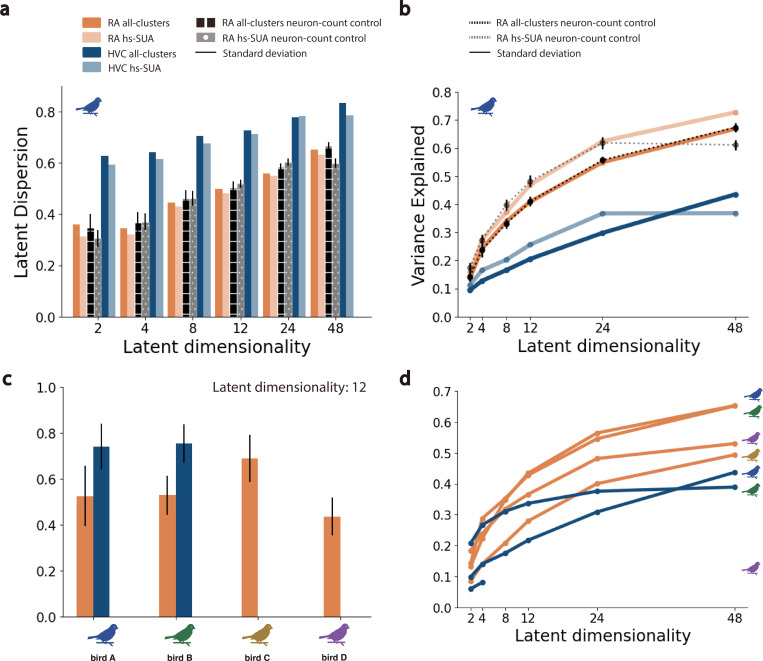
Fig. 4. Dimensionality-Dependent Analysis of Latent Neural Manifolds.
a. Mean latent dispersion, calculated as the average deviation from the mean trajectory, observed in neural manifolds inferred through GPFA as a function of latent dimensionality across different neural populations in a selected bird. Two distinct neural populations corresponding to well-isolated, high-spiking single-units (hs-SUA) and to a combination of all isolated SUA and MUA clusters (all-clusters) were considered within HVC and RA recordings. The latent dispersion for by the inferred manifolds remains stable, irrespective of the neuron type or the sorting quality within a subpopulation. Controls randomly subsampling from the RA population to match the sizes of the HVC and RA populations confirm that the resulting manifolds were robust to variations in population sizes in our recordings. b. Neural variance captured by manifolds inferred through GPFA as a function of latent dimensionality across different neural populations in a selected bird. The variance accounted for by the inferred manifolds remains consistent, irrespective of the neuron type or the sorting quality within a subpopulation. c. Latent dispersion observed in 12-dimensional latent manifolds across four different songbird individuals. d. Neural variance captured by latent manifolds across four different songbird individuals.