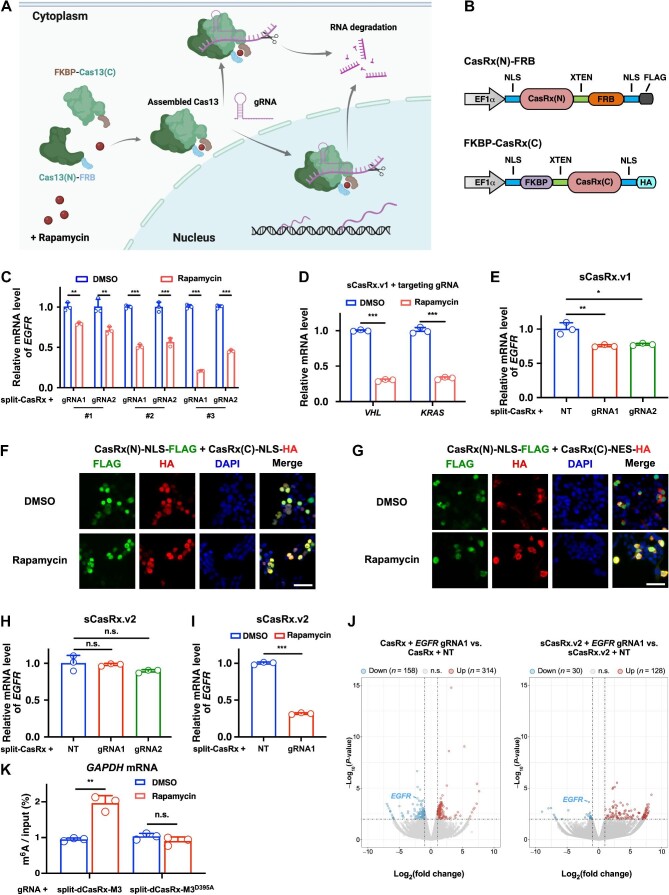
Figure 1.
Inducible RNA knockdown and m6A editing by a split-CasRx architecture. (A) Schematic illustration of the rapamycin-inducible split-Cas13 system. (B) Construction of NLS-CasRx(N)-FRB-NLS and NLS-FKBP-CasRx(C)-NLS fusion proteins (sCasRx.v1). (C) sCasRx.v1-mediated knockdown of EGFR mRNA in the presence of rapamycin. (D) sCasRx.v1-mediated knockdown of VHL and KRAS mRNAs in the presence of rapamycin. (E) sCasRx.v1-mediated knockdown of EGFR mRNA in the absence of rapamycin. (F) Cellular localization of the CasRx(N) and CasRx(C) fragments of sCasRx.v1. Scale bar, 50 μm. (G) Cellular localization of the CasRx(N) and CasRx(C) fragments of the optimized sCasRx.v2. Scale bar, 50 μm. (H) sCasRx.v2-mediated knockdown of EGFR mRNA in the absence of rapamycin. Compared with sCasRx.v1, sCasRx.v2 mediated less background RNA knockdown. (I) sCasRx.v2-mediated knockdown of EGFR mRNA in the presence of rapamycin. (J) Volcano plots showing that compared with intact CasRx, sCasRx.v2-mediated RNA knockdown affected less non-targeted mRNAs. Two biological replicates were performed. (K) Validation of rapamycin-induced RNA m6A deposition by split-dCasRx and M3 conjugates. NT, non-targeting gRNA. Values and error bars represent mean ± standard deviation. n.s., not significant; *P < 0.05; **P < 0.01; ***P < 0.001.