Figure 1: pH- and Cl−-dependent conformational rearrangements of CLC-ec1.
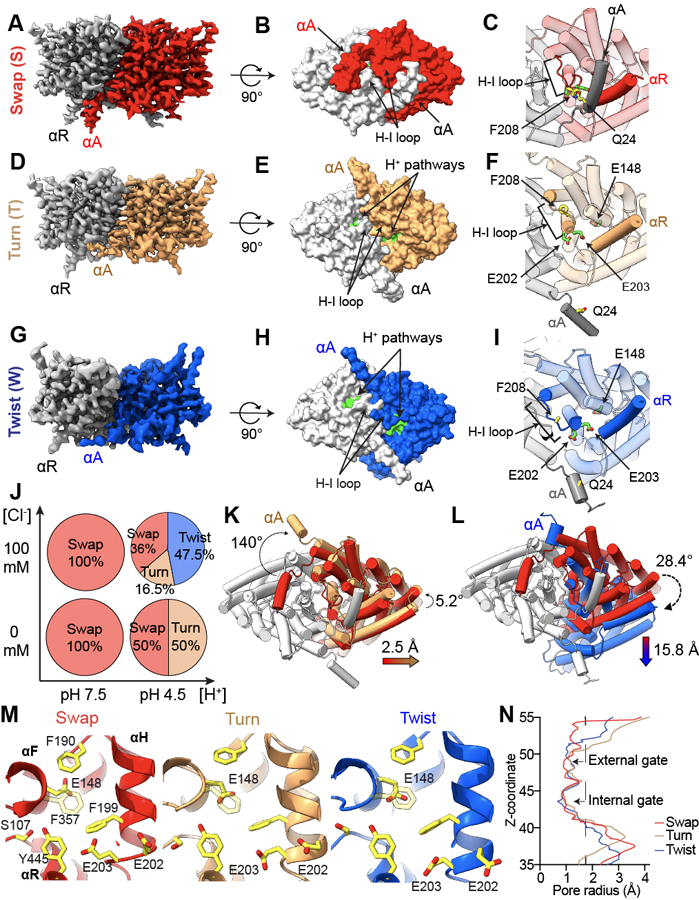
(A-I) Side views of the cryoEM density maps of CLC-ec1 in Swap (A), Turn (D), and Twist (G) imaged at pH 4.5 in 100 mM Cl−. Surface representation of CLC-ec1 in Swap (B), Turn (E), and Twist (H) viewed from the intracellular side. Residues lining the H+ pathway are colored in green (E113, E148, A182, L186, A189, F190, F199, E202, E203, M204, I402, T407 and Y445). Close-up view of gating of the H+ pathway by αA in Swap (C), Turn (F) and Twist (I). The dimers are shown in transparent cartoon representation apart from the H-I loops and helices αA and αR from opposing protomers. The H+ pathway residues E148, E202 and E203 are shown in green CPK stick representation. The interacting residues Q24 in αA and F208 in the H-I loop are shown in yellow CPK stick representation. In Twist the Coulomb density for Q24 and F208 side chains is insufficient for model building. In all panels one protomer is shown in gray and the other is colored (Swap in red, Turn in wheat, and Twist in blue). (J) A scheme showing pH and chloride conditions for cryoEM data collection, and conformational states observed in each condition: Swap (S), Turn (T), and Twist (W). The % of particles in each conformation are shown in the pie charts. (K-L) Cartoon representations of the bottom view of Turn (K) and Twist (L) aligned to Swap on protomers A. Color scheme is as in A-F. Dashed arrows denote protomer B or αA rotations, and colored arrows denote center of mass (COM) movements. (M) Close-up view of the Cl− and H+ pathways of CLC-ec1 in Swap (left), Turn (middle), and Twist (right). S107, Y445 and E148 are Cl− pathway residues, and H+ pathway residues are E148, F190, F199, E202, and E203. All residues are shown as yellow CPK sticks. (N) Pore radius profile along the z-axis of the Cl− pathway calculated using HOLE 78 for CLC-ec1 in Swap, Turn, and Twist.
