Abstract
With the increasing prevalence of age-related chronic diseases burdening healthcare systems, there is a pressing need for innovative management strategies. Our study focuses on the gut microbiota, essential for metabolic, nutritional, and immune functions, which undergoes significant changes with aging. These changes can impair intestinal function, leading to altered microbial diversity and composition that potentially influence health outcomes and disease progression. Using advanced metagenomic sequencing, we explore the potential of personalized probiotic supplements in 297 older adults by analyzing their gut microbiota. We identified distinctive Lactobacillus and Bifidobacterium signatures in the gut microbiota of older adults, revealing probiotic patterns associated with various population characteristics, microbial compositions, cognitive functions, and neuroimaging results. These insights suggest that tailored probiotic supplements, designed to match individual probiotic profile, could offer an innovative method for addressing age-related diseases and functional declines. Our findings enhance the existing evidence base for probiotic use among older adults, highlighting the opportunity to create more targeted and effective probiotic strategies. However, additional research is required to validate our results and further assess the impact of precision probiotics on aging populations. Future studies should employ longitudinal designs and larger cohorts to conclusively demonstrate the benefits of tailored probiotic treatments.
Keywords: precision probiotics, gut microbiome, older adults, metagenomics, non-negative matrix factorization, consensus clustering
Introduction
As our global population grows older, we are seeing a significant rise in age-related chronic diseases such as heart disease, cancer, diabetes, and Alzheimer’s disease, along with physical disabilities. These conditions are putting a massive strain on healthcare systems worldwide due to their increasing prevalence, costly treatments, and the need for effective management solutions and responses [1–3]. In older adults, the importance of gut microbiota becomes even more pronounced due to changes in physiology, immunity, and microbiology that accompany aging. These changes can directly affect gut health, leading to decreased taste and smell sensitivity, reduced stomach acid production, and slower digestive movement. Consequently, these alterations can impact the diversity and function of the gut microbiota, including shifts in the dominant bacterial species present [4].
Recent studies have highlighted the potential of probiotics in influencing the gut microbiota to support health and combat age-related diseases [5–7]. Probiotics are emerging as a promising strategy to rebalance the gut microbiome, boost immune function, and enhance overall health [8–10]. However, their effectiveness can vary based on several factors, such as the specific probiotic strains used and an individual’s unique gut microbiota composition, which can be influenced by lifestyle, diet, and medication use, including antibiotics [11, 12]. It is crucial to understand that the efficacy of probiotics depends on precisely matching specific strains to the right conditions and that their integration into the gut varies among individuals due to unique microbial environments [13, 14]. For example, using a probiotic strain already prevalent in an individual may lessen its beneficial impact.
In response to these challenges, we advocate for a tailored approach to probiotic supplementation, informed by a detailed understanding of an individual’s gut microbiota composition. In this study, we analyze the gut microbiota of 297 older adults with the goal of identifying probiotic signatures and exploring the gut microbiota profile, demographic, and clinical attributes associated with different probiotic signatures. This analysis is crucial for identifying probiotic strains that could effectively complement or enhance the existing gut microbiota in older adults, thereby paving the way for personalized probiotic interventions.
Materials and methods
Study design and participants
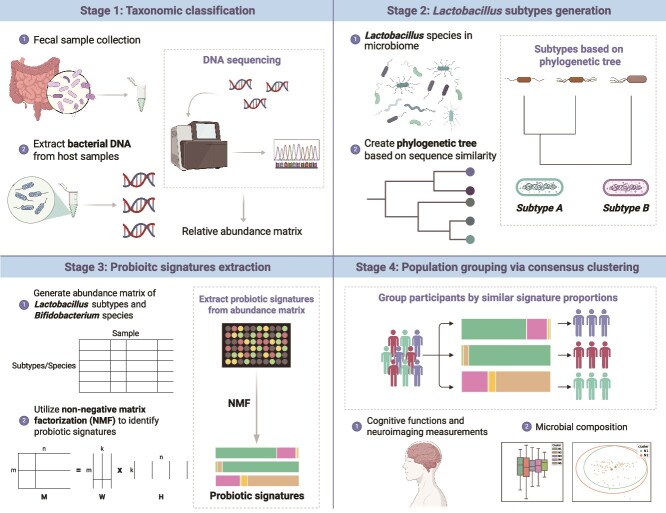
This study was conducted as part of the Taiwan Precision Medicine Initiative on Cognitive Impairment and Dementia cohort (TPMIC), a dynamic prospective cohort study aimed at establishing a biobank of blood, gut microbiota, genetic, neuroimaging, and clinical data associated with cognitive impairment. The TPMIC enrolled adults aged 50 years and above from communities and hospitals in North Taiwan, who were either cognitively normal, had mild cognitive impairment (MCI), or dementia. Participants recruited from the hospital included individuals from health checkup centers, as well as psychiatry and neurology outpatient clinics. MCI is an intermediate state between normal cognition and dementia, and the diagnosis of MCI and dementia was adjudicated by an expert panel based on NIA-AA criteria [15, 16]. Older adults with major psychiatric or neurological disorders outside of the dementia spectrum, a life expectancy of less than 6 months, recent surgery, or contraindications to MRI screening were excluded from the recruitment. This study was approved by the Far Eastern Memorial Hospital Research Ethics Committee (110065-F) and the Institutional Review Board of the Cardinal Tien Hospital (CTH-110-2-1-014). Informed consent was obtained from all participants. This study initially used 189 cognitively normal adults to establish a workflow of identifying groups of older adults with similar probiotic profiles (Fig. 1), which was then applied to 108 patients with mild dementia or MCI from August 2021 to October 2022 (Supplementary Figure S2, Table S1, Table S2, and Table S3). Participants collected a fresh fecal sample and mailed it to Allbio Biotechnology Corp. (Taichung, Taiwan). DNA was extracted using DNeasy PowerSoil Pro Kit (Qiagen, MD, USA). Sequencing and library construction of amplicon DNA samples were entrusted to AllBio Life (Taichung, Taiwan). The detailed DNA extraction and metagenomic data processing was provided in the Supplementary Material.
Figure 1.
Workflow for generating and implementing the experiment design.
Lactobacillus species detectability enhancement and subtype generation
We initially generated an abundance matrix for probiotics, including Lactobacillus and Bifidobacterium species. The low prevalence of Lactobacillus species, generally less than 5% in our dataset, challenges accurate taxonomic classification and affects the precision of the downstream analyses, particularly in individual samples. As a result, we need to increase the prevalence of Lactobacillus species by regrouping strains into subtypes. We employed standard nucleotide and amino acid similarity metrics to assess the relationships between Lactobacillus species from their whole genome sequence. At the nucleotide level, an all-against-all BLASTP alignment was executed, followed by concatenating 114 single-copy core gene families from Lactobacillaceae and Leuconostocaceae. Subsequently, we conducted phylogenetic tree analysis using the RAxML algorithm, employing the PROTGAMMAILGF (LG + I + G + F) model [17]. In the amino acid similarity level, pairwise amino acid identity and core amino acid identity were calculated to evaluate the similarity between two Lactobacillus species. Core amino acid identity was calculated from the protein sequences of the core gene families, defined as genes present in over 90% of the studied genome. We combined two similarity measures to aggregate Lactobacillus species into subtypes that reflect the summed-up abundance of member species.
Deciphering probiotic signatures by non-negative matrix factorization technique
After generating the abundance matrix for Lactobacillus subtypes and Bifidobacterium species, we used non-negative matrix factorization (NMF) to identify probiotic signatures. The NMF process starts by decomposing the probiotics abundance matrix M―comprising n probiotics species or subtypes across m samples―into two sub-matrices: the weighted matrix W (m × k) and the coefficient matrix H (k × n), each containing k components. Each component in H corresponds to a specific combination of probiotics, termed as a ‘probiotic signature’ (Supplementary Fig. S1, see online supplementary material for a color version of this figure). The weighted matrix W represents the weight of each component in samples, while the coefficient matrix H represents the composition of each component of probiotics. Each coefficient value in H quantifies the contribution or importance of a specific species within the corresponding signature. Furthermore, these coefficients were normalized by the total coefficients within the same signature to determine the relative importance of each species. Through NMF, the total abundance of sample i calculated from all signatures can be expressed as follows:
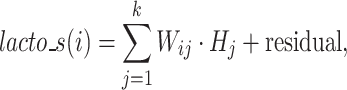 |
(1) |
where 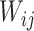 represents value of position (i, j) in weighted matrix W,
represents value of position (i, j) in weighted matrix W, 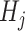 represents coefficient vector of signature j in H.
represents coefficient vector of signature j in H. 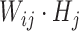 , representing the sum product of Wij and Hj, indicates the contribution of signature j to the abundance in sample i. The abundance of probiotic signature j in sample i is expressed as a percentage calculated using the following equation:
, representing the sum product of Wij and Hj, indicates the contribution of signature j to the abundance in sample i. The abundance of probiotic signature j in sample i is expressed as a percentage calculated using the following equation:
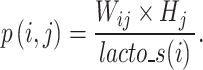 |
(2) |
Consensus clustering of probiotics profiles
We then used probiotic signatures and the consensus clustering technique to identify groups of participants with similar signature proportions. Consensus clustering is a widely used algorithm designed to determine the optimal number of clusters in a dataset. It consists of two main components: resampling and clustering. In this study, the probiotics abundance matrix, our original dataset, is randomly divided into multiple subsets, each representing 80% of the total data. Proportions of the identified signatures were calculated using the original abundance matrices of Lactobacillus subtypes and Bifidobacterium species. The matrix of signature proportion was then used as the input of the consensus clustering algorithm. Each subset then undergoes clustering for each potential number of clusters, denoted as 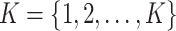 . During the clustering stage, the connectivity coefficient M and inventory coefficient I were computed to record the cluster relationship and resampling results between samples for each subset. In the subset Dh, the connectivity coefficient M of samples i and j is defined as follows:
. During the clustering stage, the connectivity coefficient M and inventory coefficient I were computed to record the cluster relationship and resampling results between samples for each subset. In the subset Dh, the connectivity coefficient M of samples i and j is defined as follows:
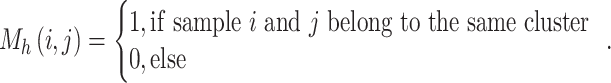 |
(3) |
The inventory coefficient I of samples i and j is defined as follows:
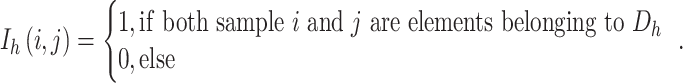 |
(4) |
Finally, the consensus coefficient C is calculated by aggregating the connectivity and inventory coefficients across all H subsets. That is, the consensus coefficient C represents a measure of the level of consensus between subsamples in terms of how often two samples are clustered together. The consensus coefficient C of the samples i and j is defined as follows:
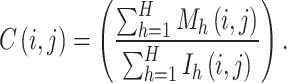 |
(5) |
After computing the consensus coefficient for all pairs of samples, the final agglomerative clustering prunes the sample into k groups called ‘consensus clusters’ based on the consensus coefficient matrix for each K cluster number. Consensus clustering was performed with ConsensusClusterPlus version 1.60.0 [18], employing hierarchical clustering based on the Pearson correlation coefficient of signature proportions between samples.
Statistical analysis of associations between probiotic consensus clusters and participant characteristics
We examined the differences between clusters in both normal and MCI/mild dementia cohorts. To investigate the dissimilarities among consensus clusters, we analyzed the correlation between probiotic clusters and various participant characteristics, including demographics, microbial composition, key microbial species, cognitive functions, and neuroimaging measurements. For more information on how these data were generated, please refer to the Supplementary Material. ANOVA was used to compare consensus clusters, followed by Bonferroni-corrected post-hoc tests for variables that showed significant results. Furthermore, we applied multiple linear regression to adjust these variables for potential confounding factors. We performed these analyses in R version 4.3.1 and Stata 18, setting the threshold for statistical significance at P < .05 for all tests.
Results
The results of the cognitively normal cohort are detailed below, with a summary of the MCI/mild dementia cohort results presented subsequently.
Relative abundance and prevalence of Lactobacillus and Bifidobacterium genera
Among 189 cognitively normal older adults, we identified 22 Lactobacillus species and 9 Bifidobacterium species. These included key probiotics like Lactobacillus casei group and Bifidobacterium longum. The analysis detailed in Supplementary Fig. S3 (see online supplementary material for a color version of this figure) illustrates the mean relative abundance and prevalence of species within the Lactobacillus and Bifidobacterium genera. Notably, five out of the nine Bifidobacterium species showed a prevalence exceeding 20%; in contrast, the highest prevalence of Lactobacillus species was merely 14.81%. Analysis of mean relative abundance revealed that more than 1% was observed in three Bifidobacterium species. Conversely, the Lactobacillus genus exhibited a significantly lower mean relative abundance, with an average below 0.1%.
Subtypes of lactobacillus species
Among the 22 Lactobacillus species identified, Lactobacillus paragasseri was assigned to the Lactobacillus subtype due to its high sequence identity (99.9%) with Lactobacillus gasseri, another species within Lactobacillus subtype. Lactobacillus rogosae was excluded from the subtypes due to its uncertain evolutionary relationship with the Lactobacillus genus [19]. As a result, 21 Lactobacillus species were organized into 8 distinct subtypes, as detailed in Table 1. The Lactobacillus subtype included the most species. When compared with the most dominant species within this subtype, there was an increase in prevalence by ~9.5%. In comparison, the Limosilactobacillus subtype, comprising four members, showed a prevalence increase of ~6.35% relative to its most dominant species. The genomic subtyping approach has effectively enhanced the identification and analysis of Lactobacillus species, leading to observed increases in their prevalence and relative abundance.
Table 1.
Prevalence and mean relative abundance of species and subtypes in the normal cohort; species under the subtype is the members of subtype
| Prevalence (%) | Mean relative abundance (%) | |
|---|---|---|
| Lactobacillus subtype | 15.87 | 4.65E−02 |
| Lactobacillus acidophilus | 4.23 | 6.64E−03 |
| Lactobacillus amylovorus | 1.59 | 1.27E−02 |
| Lactobacillus crispatus | 2.65 | 1.36E−03 |
| Lactobacillus delbrueckii | 6.35 | 2.47E−02 |
| Lactobacillus gasseri | 2.65 | 6.28E−04 |
| Lactobacillus iners | 0.53 | 3.37E−05 |
| Lactobacillus johnsonii | 0.53 | 3.72E−04 |
| Lactobacillus paragasseri | 2.65 | 1.18E−04 |
| Ligilactobacillus subtype | 15.87 | 4.95E−02 |
| Lactobacillus ruminis | 1.06 | 4.68E−03 |
| Lactobacillus salivarius | 14.81 | 4.48E−02 |
| Lacticaseibacillus subtype | 6.35 | 9.81E−03 |
| Lactobacillus casei group | 4.23 | 8.76E−03 |
| Lactobacillus rhamnosus | 3.17 | 1.05E−03 |
| Latilactobacillus subtype | 1.06 | 8.36E−04 |
| Lactobacillus curvatus | 0.53 | 3.29E−04 |
| Lactobacillus sakei | 0.53 | 5.07E−04 |
| Companilactobacillus subtype | 1.06 | 5.21E−04 |
| Lactobacillus farciminis | 1.06 | 5.21E−04 |
| Limosilactobacillus subtype | 13.23 | 2.79E−02 |
| Lactobacillus fermentum | 6.88 | 1.41E−02 |
| Lactobacillus mucosae | 6.88 | 1.32E−02 |
| Lactobacillus oris | 1.59 | 1.45E−04 |
| Lactobacillus vaginalis | 3.17 | 4.02E−04 |
| Lactiplantibacillus subtype | 6.35 | 9.30E−03 |
| Lactobacillus plantarum | 6.35 | 9.30E−03 |
| Fructilactobacillus subtype | 0.53 | 2.13E−04 |
| Lactobacillus sanfranciscensis | 0.53 | 2.13E−04 |
Probiotic signatures and consensus clusters
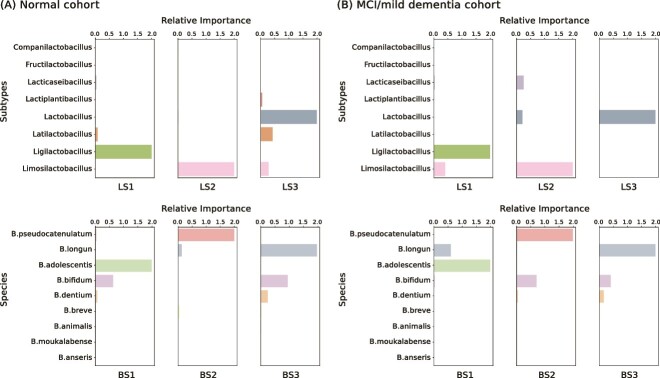
Based on an analysis of 189 cognitively normal samples, we have identified three signatures each for Lactobacillus and Bifidobacterium. These signatures, detailed in Fig. 2A, emphasize their relative importance to overall probiotic signature. The first Lactobacillus Signature (LS1) is characterized by a strong presence of the Ligilactobacillus subtype, which includes species such as Lactobacillus ruminis and Lactobacillus salivarius. LS2 is dominated by the Limosilactobacillus subtype, including well-documented probiotics such as Lactobacillus fermentum. LS3 comprises a complex blend of Lactobacillus, Latilactobacillus, and Limosilactobacillus subtypes. Within the Bifidobacterium Signatures (BS), BS1 combines Bifidobacterium adolescentis and Bifidobacterium bifidum; BS2 prominently features Bifidobacterium pseudocatenulatum; while BS3 is a composite of B. longum, B. bifidum, and Bifidobacterium dentium, with relative importance values of 1.96, 0.94, and 0.25, respectively. We successfully identified these probiotic signatures using NMF.
Figure 2.
Relative importance of Lactobacillus subtypes in three LS and relative importance of Bifidobacterium species in three BS, presenting the components of each probiotic signature; (A) relative importance in the normal cohort; (B) relative importance in the MCI/mild dementia cohort.
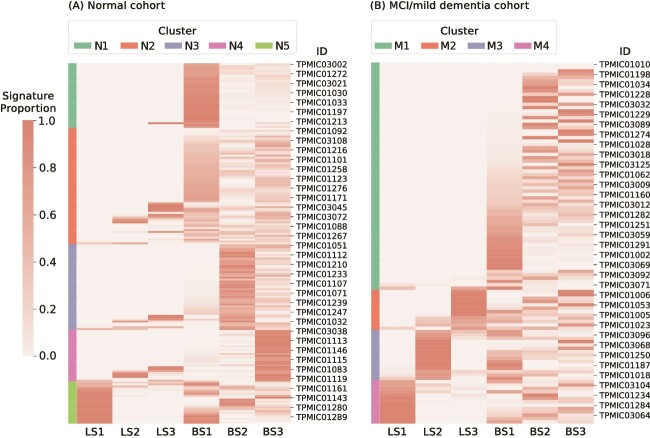
After entering the signature proportion matrix into the clustering algorithm, we categorized the 189 samples into five distinct clusters, as shown in Fig. 3A. Notably, samples in clusters N1, N3, and N4 predominantly featured BS1, BS2, and BS3, respectively. Samples in cluster N5 were enriched with LS1 and BS1; while samples in cluster N2 displayed a diverse combination of probiotic signatures, indicating varied probiotic profiles.
Figure 3.
Signature proportion heatmap of (A) the normal cohort and (B) the MCI/mild dementia cohort, and consensus clustering was done based on the probiotic signature proportion matrix; the color bar represents the results of consensus clusters.
Gut microbial, demographic, cognitive, and brain characteristics among consensus clusters
We analyzed microbial composition, demographic characteristics, cognitive functions, and brain structures to elucidate differences among clusters within the cognitively normal cohort. Our analysis revealed only a few differences in the alpha diversity of the entire microbiome, as shown in Fig. 4A. Only clusters N3 and N4, compared with cluster N5, showed notable differences in both Chao1 and Fisher indices. The beta diversity analysis further highlighted significant variations between cluster N4 when compared with clusters N1 and N2, as determined by the PERMANOVA test (Fig. 4B). These findings demonstrate the potential role of probiotic composition variations in shaping the gut microbiome ecosystem. We employed the LEfSe (Linear discriminant analysis Effect Size) method to identify characteristic species within each cluster, highlighting those with statistically significant differences. Several species exhibited differentiated abundance across five clusters. To name a few, Prevotella copri, Streptococcus sobrinus, and Fusobacterium varium are the representative species in cluster N2. Ruminococcus gnavus, another non-probiotic species, was found to be predominant in cluster N4. More key species were linked to cluster N5, including Bacteroides plebeius, Megamonas funiformis, Streptococcus salivarius, Enterobacter mori, Streptococcus parasanguinis, Scardovia wiggsiae, and Tyzzerella spp. (Fig. 4C).
Figure 4.
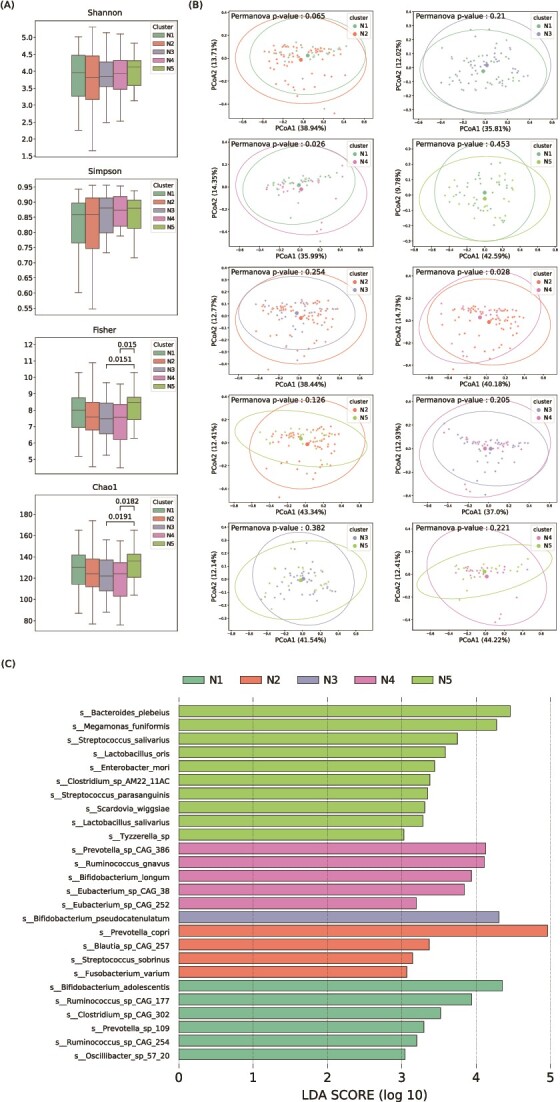
Differences in microbial composition between clusters in the normal cohort; (A) Shannon, Simpson, fisher, and Chao1 are alpha diversity indices; the Wilcoxon rank-sum tests were used to compare the differences, and only significant differences (P-value < .05) were displayed; (B) PERMAVONA tests were conducted to display the differences in beta diversity, and each point represents the coordinates of a sample in vector space after PCoA transformation based on the weighted-Unifrac distance matrix, and (C) representative species in each consensus cluster; the x-axis means species log-transformed LDA score from LEfSe, which represents the effect size of species; y-axis is the list of candidate species.
Table 2 details the demographic characteristics, cognitive assessments, and brain structure indices across five consensus clusters, providing insights beyond microbiome-driven distinctions. Clusters N1 and N3 mainly consisted of younger individuals with higher education, in contrast to cluster N5, which mainly comprised older men with lower education levels. Although no significant differences were found in age, gender, or education levels among the clusters after performing ANOVA, a pattern emerged concerning smoking and drinking habits. Specifically, clusters N2 and N5 had a higher percentage of smokers and drinkers, while cluster N1 individuals predominantly refrained from both. Following Bonferroni corrections, the post-hoc analysis showed a significant difference in drinking habits: only 10% of individuals in cluster N1 reported drinking compared with 52% in cluster N5.
Table 2.
Analysis of variance of characteristics, neuropsychological tests scores, and brain structure between consensus clusters in the normal cohort
| Characteristics | Cluster N1 n = 34 |
Cluster N2 n = 61 |
Cluster N3 n = 45 |
Cluster N4 n = 27 |
Cluster N5 n = 22 |
P-value |
|---|---|---|---|---|---|---|
| Age, years, mean ± SD | 66.6 ± 7.1 | 67.5 ± 6.7 | 66.5 ± 6.1 | 69.5 ± 6.6 | 69.6 ± 7.3 | .18 |
| Male, N(%) | 13 (38) | 22 (36) | 16 (36) | 7 (26) | 14 (64) | .091 |
| Education years, mean ± SD | 13.1 ± 3.4 | 12.1 ± 4.4 | 13.3 ± 3.6 | 11.3 ± 3.4 | 11.2 ± 3.9 | .080 |
| Smoking ever, N(%) | 2 (7) | 15 (26) | 6 (14) | 2 (8) | 7 (33) | .034 |
| Drinking ever, N(%) | 3 (10)a | 21 (36) | 14 (33) | 5 (20) | 11 (52)a | .011 |
| BMI, mean ± SD | 24.6 ± 3.8 | 23.9 ± 3.8 | 24.7 ± 4.1 | 23.4 ± 3.6 | 24.7 ± 5.1 | .57 |
| Diabetes mellitus, N(%) | 5 (17) | 10 (17) | 12 (29) | 3 (12) | 4 (19) | .48 |
| Hypertension, N(%) | 9 (30) | 24 (42) | 16 (38) | 9 (36) | 8 (38) | .87 |
| Score, mean ± SD | n = 34 | n = 61 | n = 45 | n = 27 | n = 22 | P-value |
| MMSE | 27.8 ± 2.1 | 28.4 ± 1.7 | 27.9 ± 2.3 | 28.7 ± 1.4 | 28.4 ± 2.0 | .27 |
| LMII | 21.5 ± 5.9 | 20.2 ± 6.9 | 22.2 ± 8.2 | 22.7 ± 6.3 | 19.6 ± 7.8 | .34 |
| LMI | 37.4 ± 8.1 | 35.1 ± 9.7 | 38.1 ± 9.4 | 37.6 ± 8.0 | 33.2 ± 12.3 | .22 |
| DS | 28.3 ± 4.4 | 26.7 ± 5.7 | 26.2 ± 5.7 | 27.2 ± 5.6 | 26.5 ± 4.3 | .54 |
| DSST | 66.2 ± 14.0 | 61.2 ± 16.6 | 70.2 ± 17.0aa | 58.7 ± 18.2a | 54.9 ± 16.5a | .002 |
| CTT1 (s) | 47.1 ± 15.4 | 53.1 ± 18.0 | 57.6 ± 33.8 | 60.6 ± 31.8 | 57.9 ± 19.9 | .20 |
| VF | 39.4 ± 6.7a | 38.7 ± 8.5 | 37.9 ± 7.0 | 37.3 ± 6.2 | 33.5 ± 7.5a | .043 |
| CTT2 (s) | 105.1 ± 33.9 | 114.8 ± 39.4 | 109.2 ± 48.2 | 135.6 ± 55.9 | 129.8 ± 45.6 | .035 |
| SCWT-interference | 78.2 ± 14.8 | 79.1 ± 15.2 | 80.0 ± 17.7 | 78.4 ± 17.6 | 70.3 ± 12.0 | .18 |
| Memory | 0.04 ± 0.83 | −0.14 ± 0.97 | 0.14 ± 1.16 | 0.22 ± 0.89 | −0.22 ± 1.10 | .34 |
| Attention | 0.21 ± 0.49 | −0.05 ± 0.75 | 0.09 ± 0.81 | −0.08 ± 0.73 | −0.25 ± 0.77 | .15 |
| Executive function | 0.17 ± 0.51a | 0.01 ± 0.66 | 0.06 ± 0.61 | −0.06 ± 0.77 | −0.34 ± 0.69a | .065 |
| Volume, cm3, mean ± SD | n = 28 | n = 57 | n = 45 | n = 27 | n = 21 | P-value |
| Total brain volume | 1072.2 ± 87.9 | 1052.5 ± 92.9 | 1053.1 ± 93.3 | 1042.3 ± 91.4 | 1086.1 ± 90.3 | .44 |
| Ventricle | 21.4 ± 6.1aa | 30.0 ± 14.7a | 28.8 ± 13.2 | 28.3 ± 11.5 | 33.9 ± 12.5a | .010 |
| Cortex | 433.3 ± 34.8 | 430.8 ± 36.0 | 432.2 ± 33.6 | 425.5 ± 36.7 | 438.8 ± 36.7 | .78 |
| Gray matter | 594.0 ± 44.7 | 590.2 ± 46.7 | 591.1 ± 42.4 | 581.1 ± 47.2 | 604.8 ± 47.0 | .50 |
| White matter | 450.6 ± 50.0 | 435.2 ± 49.8 | 435.0 ± 53.2 | 435.8 ± 49.1 | 454.6 ± 49.7 | .39 |
| Hippocampus | 8.4 ± 0.9 | 8.0 ± 0.8 | 8.1 ± 0.7 | 7.8 ± 0.8 | 8.0 ± 1.0 | .16 |
| Entorhinal | 3.8 ± 0.5 | 3.7 ± 0.6 | 3.6 ± 0.6 | 3.7 ± 0.7 | 3.6 ± 0.6 | .90 |
| Amygdala | 3.3 ± 0.4a | 3.1 ± 0.4 | 3.1 ± 0.3 | 3.0 ± 0.3a | 3.1 ± 0.4 | .037 |
| Thickness, mm, mean ± SD | n = 28 | n = 57 | n = 45 | n = 27 | n = 21 | P-value |
| AD-score | 2.53 ± 0.11 | 2.55 ± 0.11a | 2.52 ± 0.08 | 2.51 ± 0.07 | 2.47 ± 0.10a | .025 |
BMI, body max index; LM, Logical Memory Test; DS, Digit Span; VF, Semantic Verbal Fluency; SCWT, Stroop Color and Word Test.
aThere is a significant difference (P-value < .05) between groups after ANOVA post-hoc tests under Bonferroni correction.
Cognitive assessments revealed significant differences among clusters in Digit Symbol Substitute Test (DSST), Semantic Verbal Fluency (VF), and Color Trails Test 2 (CTT2) scores, with the executive function domain demonstrating larger heterogeneity. Older adults in cluster N5 scored the lowest on the DSST and VF tests when compared with clusters N3 and N1, respectively. Brain imaging revealed that participants in clusters N2 and N5 had enlarged ventricles compared with those in cluster N1. Additionally, the AD signature cortices were the thinnest in cluster N5 and the thickest in cluster N2. Diffusion Tensor Imaging (DTI) analysis highlighted significant variances in the bilateral fornix, right hippocampus, and right tapetum, with FA values being lowest in cluster N5 and highest in cluster N1 (Supplementary Table S4, see online supplementary material for a color version of this table). These results emphasized the consensus clustering’s efficacy in distinguishing other characteristics among older adults. However, multiple linear regression analyses, accounting for confounders as shown in Table 3, did not yield statistically significant differences. After adjustment, only differences in drinking habits and amygdala volume remained significantly varied across clusters.
Table 3.
Multiple regression analysis of characteristics different between consensus clusters after adjusting for potential confounding factors in the normal cohort
| P-value | |
|---|---|
| Smoking evera | .094 |
| Drinking evera | .030 |
| DSSTa | .18 |
| VFa | .19 |
| CTT2 (sec)a | .49 |
| Executive functiona | .27 |
| Ventricleb | .059 |
| Amygdalab | .029 |
| AD-scoreb | .078 |
| Cingulum (hippocampus) Rb,c | .089 |
| Fornixb,d | .28 |
| Cingulum (hippocampus) Rb,d | .14 |
| Fornix (cres)/Stria terminalis Rb,d | .050 |
| Fornix (cres)/Stria terminalis Lb,d | .19 |
| Tapetum Rb,d | .073 |
aadjusted for age, gender, and years of education.
badjusted for age, gender, and estimated total intracranial volume.
cJHU WM tractography atlas.
dICBM-DTI-81 WM labels atlas.
Results of the MCI/mild dementia cohort
Applying the same workflow of analysis (Fig. 1) in 108 MCI/mild dementia patients, we also identified distinct probiotic signatures and clustering patterns. First, we identified 26 Lactobacillus species and 9 Bifidobacterium species, and the Lactobacillus species were also organized into 8 distinct subtypes. We then discerned three distinctive LSs and three BSs. LS1, LS2, and LS3 were characterized by the predominance of Ligilactobacillus, Limosilactobacillus, and Lactobacillus subtypes, respectively, detailed in Fig. 2B. Similarly, B. adolescentis, B. pseudocatenulatum, and B. longum were identified as the principal components of BS1, BS2, and BS3.
Contrasting with the cognitively normal group, the MCI cohort was categorized into four unique clusters based primarily on their LS, employing a consensus clustering technique. Cluster M1 was predominantly characterized by BS, showing an absence of LS. In cluster M2, a notable association with LS3 was noted, whereas LS2 and LS1 were the defining features of clusters M3 and M4, respectively, as demonstrated in Fig. 3B.
For gut microbial analysis, the alpha diversity revealed significant differences in Fisher and Chao1 indices of clusters M1 and M3 (Supplementary Fig. S4, see online supplementary material for a color version of this figure). Beta diversity analysis showed no significant differences among the clusters. Unlike the normal cohort, the MCI cohort revealed a fewer number of characteristic species. For example, Bacteroides uniformis, Clostridium asparagiforme, and Oxalobacter formigenes served as hallmark species in cluster M2. Atopobium parvulum and Allisonella histaminiformans were more abundant in cluster M4. Table 4 delineates the demographic characteristics, neuropsychological tests, and brain structure variances across four consensus clusters. However, limited sample size resulted in significant differences only between clusters M1 and M2 in CTT1 scores. Right corticospinal tract, left medial lemniscus, and left superior corona radiata were different in FA values among clusters (Supplementary Table S5, see online supplementary material for a color version of this table).
Table 4.
Analysis of variance of characteristics, neuropsychological tests scores, and brain structure between consensus clusters in the MCI cohort
| Characteristics | Cluster M1 n = 68 |
Cluster M2 n = 12 |
Cluster M3 n = 15 |
Cluster M4 n = 13 |
P-value |
|---|---|---|---|---|---|
| Age, years, mean ± SD | 73.5 ± 7.4 | 73.2 ± 8.8 | 75.5 ± 9.2 | 73.9 ± 5.5 | .83 |
| Male, N(%) | 26 (38) | 3 (25) | 7 (47) | 6 (46) | .65 |
| Education years, mean ± SD | 10.3 ± 4.3 | 7.4 ± 5.0 | 8.1 ± 4.0 | 8.5 ± 3.8 | .059 |
| Smoking ever, N(%) | 15 (23) | 4 (33) | 0 (0) | 4 (31) | .14 |
| Drinking ever, N(%) | 15 (23) | 4 (33) | 4 (29) | 3 (23) | .86 |
| BMI, mean ± SD | 23.3 ± 4.0 | 24.9 ± 4.3 | 25.1 ± 3.8 | 24.9 ± 3.6 | .24 |
| Diabetes mellitus, N(%) | 15 (23) | 4 (33) | 3 (21) | 4 (31) | .83 |
| Hypertension, N(%) | 25 (38) | 4 (33) | 6 (43) | 6 (46) | .91 |
| Score, mean ± SD | n = 68 | n = 12 | n = 15 | n = 13 | P-value |
| MMSE | 21.4 ± 6.7 | 19.7 ± 7.0 | 20.5 ± 6.1 | 19.7 ± 6.5 | .73 |
| LMII | 8.0 ± 7.8 | 4.3 ± 4.7 | 7.2 ± 6.2 | 5.5 ± 5.4 | .44 |
| LMI | 17.9 ± 10.5 | 13.9 ± 7.5 | 17.5 ± 8.3 | 15.8 ± 10.2 | .69 |
| DS | 21.2 ± 5.0 | 20.3 ± 8.0 | 17.4 ± 6.0 | 19.0 ± 4.8 | .17 |
| DSST | 41.2 ± 18.6 | 34.9 ± 27.9 | 38.0 ± 16.8 | 36.2 ± 18.1 | .73 |
| CTT1 (sec) | 101.3 ± 56.1a | 180.2 ± 139.8a | 119.4 ± 90.6 | 94.6 ± 59.1 | .027 |
| VF | 24.4 ± 9.3 | 19.2 ± 5.4 | 23.1 ± 7.5 | 18.8 ± 4.0 | .088 |
| CTT2 (sec) | 224.9 ± 96.5 | 220.3 ± 106.8 | 193.9 ± 92.1 | 192.0 ± 78.6 | .63 |
| SCWT-interference | −11.3 ± 10.0 | −7.1 ± 5.0 | −3.3 ± 13.9 | −7.7 ± 12.6 | .14 |
| Memory | 0.11 ± 1.10 | −0.40 ± 0.66 | −0.00 ± 0.87 | −0.23 ± 0.76 | .44 |
| Attention | 0.13 ± 0.74 | −0.37 ± 1.27 | −0.19 ± 0.64 | −0.08 ± 0.71 | .26 |
| Executive function | −0.09 ± 0.78 | −0.17 ± 0.62 | 0.23 ± 0.69 | −0.10 ± 0.60 | .50 |
| Volume, cm3, mean ± SD | n = 60 | n = 10 | n = 12 | n = 9 | P-value |
| Total brain volume | 1003.5 ± 101.3 | 974.4 ± 91.1 | 991.2 ± 116.4 | 994.9 ± 68.0 | .85 |
| Ventricle | 42.7 ± 20.2 | 37.5 ± 12.7 | 46.8 ± 23.5 | 41.1 ± 10.2 | .72 |
| Cortex | 404.1 ± 43.9 | 401.6 ± 40.3 | 395.4 ± 38.1 | 408.5 ± 40.9 | .90 |
| Gray matter | 557.0 ± 55.6 | 552.5 ± 45.5 | 548.1 ± 48.7 | 556.6 ± 47.1 | .96 |
| White matter | 421.6 ± 54.3 | 399.7 ± 49.4 | 417.8 ± 65.7 | 415.9 ± 32.3 | .70 |
| Hippocampus | 7.0 ± 1.2 | 7.0 ± 1.0 | 7.0 ± 1.5 | 6.9 ± 0.9 | 1.00 |
| Entorhinal | 3.1 ± 0.9 | 3.4 ± 1.2 | 2.8 ± 0.8 | 3.3 ± 0.7 | .33 |
| Amygdala | 2.7 ± 0.5 | 2.6 ± 0.6 | 2.6 ± 0.7 | 2.6 ± 0.5 | .94 |
| Thickness, mm, mean ± SD | n = 60 | n = 10 | n = 12 | n = 9 | P-value |
| AD-score | 2.45 ± 0.13 | 2.43 ± 0.11 | 2.45 ± 0.05 | 2.48 ± 0.09 | .84 |
aThere is a significant difference (P-value < .05) between groups after ANOVA post-hoc tests under Bonferroni correction.
Discussion
This study advanced our understanding of population clustering through the lens of probiotics species, specifically Lactobacillus and Bifidobacterium, in older adults. Notably, there are distinct demographic characteristics, lifestyle habits, cognitive performance, and brain structure variance among the clusters. Our methodology was demonstrated in both cognitively normal and MCI/mild dementia cohorts. Probiotic profiles in the MCI/mild dementia cohort significantly differed from those in the cognitively normal group, indicating that our methodology shows potential to identify phenotype-specific traits embedded within the samples. Our findings advocate for a holistic approach to understanding the gut-brain axis, emphasizing the importance of considering both probiotics microbiome composition and broader health indicators in cognitive health research.
The concept of ‘probiotic patterns’ and ‘probiotic clusters’ remain relatively unexplored in the current scientific literature. The pioneering work by Zhang et al. and Ghosh et al. has begun to shed light on this area. Zhang et al. utilized heatmap visualization and an unsupervised K-means clustering algorithm to categorize 40 Lactobacillus strains based on their probiotic attributes [20]. Ghosh et al. applied a similar clustering approach to discern 25 Lactobacillus species of varying prevalence within the microbiomes of ostensibly healthy individuals. Their analysis identified six distinct ‘Lactobacillotypes’: three dominated by Lactobacillus delbrueckii, L. ruminis, and Lacticaseibacillus casei, respectively, with the remaining three representing a mix of various species. Notably, the predominant species in Ghosh et al.’s study align with the Lactobacillus, Ligilactobacillus, and Lacticaseibacillus subtypes delineated in our research. These classifications unveil unique, age-specific microbiotic patterns influenced by geographical location and correlating with demographic and health-related factors such as age, gender, BMI, and disease prevalence [21].
Although the development of probiotic clusters occurred independently of other sample characteristics or phenotypes, integrating the demographic, cognitive, and neuroimaging information revealed distinct probiotic compositions and characteristics among consensus clusters. These findings align with and extend upon findings from previous research, indicating a significant role of specific probiotic strains in mental health and aging. For example, the contrast between clusters N1 and N5 is most evident; members in the cluster N1 are younger, more educated, and lead healthier lifestyles. Bifidobacterium adolescentis, identified predominantly in cluster N1, is noted for its production of gamma-aminobutyric acid (GABA), a neurotransmitter implicated in modulating the gut microbiome and mental health disorders such as depression and anxiety [22, 23]. Studies utilizing mouse models have shown that B. adolescentis exerts anxiolytic and antidepressant effects. These effects are achieved by rebalancing of gut microbiota, reducing inflammatory cytokines such as interleukin-1β (IL-1β), tumor necrosis factor α (TNF-α), and p-nuclear factor-kappa B, and increasing brain-derived neurotrophic factor (BDNF) expression within the hippocampus [24]. Specifically, dietary supplementation with B. adolescentis has been shown to ameliorate osteoporosis and neurodegeneration in models of premature aging, with observations indicating diminished levels of this probiotic in individuals over 60 [25]. In contrast, cluster N5’s microbiota, characterized by the presence of L. ruminis (LS1), has been implicated in the brain’s inflammatory processes. A study within a Japanese stroke cohort identified a positive correlation between increased levels of L. ruminis and ischemic stroke, alongside interleukin-6 (IL-6) levels [26, 27]. Lactobacillus ruminis has been proposed as a potential biomarker for ultra-high-risk psychosis due to its tumor necrosis factor (TNF)-stimulating properties [28, 29].
In cluster N3, defined by the predominance of B. pseudocatenulatum (BS2), notable findings highlight its beneficial impact on cognitive health. Research indicates that individuals with normal cognitive function exhibit a higher abundance of B. pseudocatenulatum compared with those with dementia. This bacterium’s relative abundance is positively associated with key cognitive assessment scores, including the Mini-Mental State Examination (MMSE) and both short and long delay recall tests [30]. This correlation suggests a potential protective role of B. pseudocatenulatum against cognitive decline. Additionally, B. pseudocatenulatum levels tend to decrease with age and are negative correlated with several cytokines involved in inflammatory processes, such as IL-6, IL-1β, IL-17, TNF-α, IL-12, and TGF-β, as well as Amyloid-β (Aβ), a marker associated with Alzheimer’s disease. Conversely, its abundance positively correlates with BDNF levels, further indicating its capacity to support neural and immune system health [31]. The cluster N4, characterized by the presence of B. longum (BS3), has been shown to mitigate depression and modulate responses to negative emotional stimuli, particularly in the amygdala and fronto-limbic regions of patients with irritable bowel syndrome [32]. Additionally, R. gnavus exhibited a significant positive correlation with the Depression Anxiety Stress Scales, suggesting its influence on emotional health [33]. Our findings are consistent with previous studies, underscoring the potential complex interactions between specific probiotic strains, other microbes, and mental health.
The clusters developed from our method may be used to design precision probiotic strategies. Probiotics supplement regimens can be tailored based on individuals’ cluster groups, and future trials using this strategy can further help confirm the specific effects or mechanisms of probiotics on improving health outcomes or diseases. This approach aligns with the top-down approach of precision probiotics development, which involves identifying strains associated with specific health conditions based on observational evidence and then testing their effectiveness in animal models and humans [34, 35]. Other methods, such as genome-wide association studies and transcriptome-wide association studies (TWAS), can be valuable in precision probiotics research by examining the genes of the host or gut microbiota. Pan et al. conducted a TWAS using host genome data to associate host gene expression with gut microbiota traits [36]. Alternatively, Zahavi et al. focused on the microbial genome and conducted metagenome-wide association studies to identify connections between bacterial SNPs and human traits, suggesting potential mechanistic links with host phenotypes [37]. Regardless of the approach, developing algorithms that match person-specific data and factors influencing probiotic efficacy is crucial for identifying the optimal probiotic modality for different populations [34].
Our study presents significant strengths, notably its application to a well-characterized community-based cohort study that includes a broad spectrum of data on older adults, from cognitive performance to brain imaging. Utilizing shotgun metagenomic sequencing and advanced analytical pipelines has allowed for highly accurate assessments of the microbiome’s taxonomic composition. Moreover, the application of our clustering methodologies across both cognitively normal individuals and those with MCI underscores the potential of our approach for developing targeted probiotic supplements for cognitive health. Nonetheless, the study faces limitations. The single collection point of samples limits our ability to draw causal conclusions, compounded by a lack of data on prior antibiotic or probiotic use and metabolic profiles, which may be unmeasured confounding factors. Additionally, the absence of comprehensive diet information and metabolome data in this study means that we can only postulate potential mechanisms, guided by previous studies, rather than direct inference. Furthermore, socioeconomic factors, known to affect cognitive health, may also influence individuals’ diet choices, lifestyles, behaviors, overall health status, and consequently, the gut microbiota. Residual confounding may persist in our study as we only adjusted for educational attainment in our association analyses. Although our findings provide valuable insights for older adults in Taiwan, their generalizability to diverse global populations remains uncertain. While the sample size is adequate, it may not be sufficient to definitively establish probiotic signatures and consensus clusters, underscoring the need for expanded future research.
In conclusion, this study effectively classified older adults into distinct clusters by analyzing the signatures of Lactobacillus and Bifidobacterium derived from the microbiome abundance matrix. This classification, applicable to both cognitively normal and MCI cohorts, highlighted how unique probiotic profiles correlate with variations in population attributes, microbial compositions, cognitive functions, and neuroimaging outcomes. These findings emphasize the potential of customized probiotic interventions to enhance cognitive health. Validating the durability and effectiveness of this clustering approach requires ongoing longitudinal monitoring of the cohort.
Key Points
Advanced metagenomic sequencing was utilized to examine the gut microbiota of 297 older adults, underscoring the significance of gut microbiota in metabolic, nutritional, and immune functions, which are notably affected by aging.
We used Lactobacillus and Bifidobacterium to identify clusters of older adults with distinct probiotic signatures, and there are differences in demographic characteristics, lifestyles, cognitive functions, and brain structures among clusters.
Potential implications include the feasibility of personalized probiotic supplements to address age-related diseases and functional declines.
Supplementary Material
Acknowledgements
We appreciate all participants’ contributions to the Taiwan Precision Medicine Initiative on Cognitive Impairment and Dementia cohort (TPMIC) study. The authors acknowledge the ‘technical services’ provided by the National Genomics Center for Clinical and Biotechnological Applications of the Cancer Progression Research Center (National Yang Ming Chiao Tung University). The Core facility is supported by National Core Facility for Biopharmaceuticals (NCFB), National Science and Technology Council.
Conflict of interest: None declared.
Contributor Information
Yi-Fang Chuang, School of Medicine, National Yang Ming Chiao Tung University, No. 155, Sec. 2, Linong St. Beitou Dist., Taipei City 112304, Taiwan; Institute of Public Health, National Yang Ming Chiao Tung University, No. 155, Sec. 2, Linong St. Beitou Dist., Taipei City 112304, Taiwan; Department of Psychiatry, Far Eastern Memorial Hospital, No. 21, Sec. 2, Nanya S. Rd., Banqiao Dist., New Taipei City 220216, Taiwan.
Kang-Chen Fan, School of Medicine, National Yang Ming Chiao Tung University, No. 155, Sec. 2, Linong St. Beitou Dist., Taipei City 112304, Taiwan.
Yin-Yuan Su, Institute of Biomedical Informatics, National Yang Ming Chiao Tung University, No. 155, Sec. 2, Linong St. Beitou Dist., Taipei City 112304, Taiwan.
Ming-Fong Wu, Institute of Biomedical Informatics, National Yang Ming Chiao Tung University, No. 155, Sec. 2, Linong St. Beitou Dist., Taipei City 112304, Taiwan.
Yen-Ling Chiu, Department of Medical Research, Far Eastern Memorial Hospital, No. 21, Sec. 2, Nanya S. Rd., Banqiao Dist., New Taipei City 220216, Taiwan; Graduate Program in Biomedical Informatics and Graduate Institute of Medicine, Yuan Ze University, No. 135, Yuandong Rd., Zhongli Dist., Taoyuan City 320315, Taiwan; Graduate Institute of Clinical Medicine, National Taiwan University, No. 1, Sec. 4, Roosevelt Rd., Taipei City 106319, Taiwan.
Yi-Chien Liu, Department of Neurology, Cardinal Tien Hospital, No. 362, Zhongzheng Rd., Xindian Dist., New Taipei City 231009, Taiwan.
Chen-Ching Lin, Institute of Biomedical Informatics, National Yang Ming Chiao Tung University, No. 155, Sec. 2, Linong St. Beitou Dist., Taipei City 112304, Taiwan.
Funding
This study was supported by Taiwan National Science and Technology Council, previously Ministry of Science and Technology (MOST 110-2321-B-418, NSTC 113-2321-B-418-003, 113-2321-B-418-005, 109-2221-E-010-014, and 112-2221-E-A49-106), Ministry of Health and Welfare (MOHW111-TDU-B-221-114007 and MOHW112-TDU-B-221-124007), and FEMH-NYCU Joint Research Program (#NYCU-FEMH 112DN11).
Data availability
The relative abundance matrix of older adults’ gut microbiome composition has been provided as supplementary materials. The raw sequencing data underlying this article cannot be shared publicly due to the privacy of individuals that participated in the study. The data will be shared on the request to the first and corresponding author.
Author contributions
Conceptualization: C.C.L., Y.F.C., Y.Y.S.
Methodology: C.C.L., K.C.F., M.F.W., Y.F.C., Y.Y.S.
Data collection: Y.F.C., Y.C.L.
Implementation: K.C.F., M.F.W.
Supervision: C.C.L., Y.F.C., Y.L.C.
Writing—original draft: K.C.F., M.F.W.
Writing—review & editing: C.C.L., K.C.F., Y.F.C.
References
- 1. Fung TC, Olson CA, Hsiao EY. Interactions between the microbiota, immune and nervous systems in health and disease. Nat Neurosci 2017;20:145–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Valdes AM, Walter J, Segal E. et al. . Role of the gut microbiota in nutrition and health. BMJ 2018;361:36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Morais LH, Schreiber HL, Mazmanian SK. The gut microbiota–brain axis in behaviour and brain disorders. Nat Rev Microbiol 2020;19:241–55. [DOI] [PubMed] [Google Scholar]
- 4. Salazar N, Valdés-Varela L, González S. et al. . Nutrition and the gut microbiome in the elderly. Gut Microbes 2017;8:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hamilton-Miller JMT. Probiotics and prebiotics in the elderly. Postgrad Med J 2004;80:447–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ale EC, Binetti AG. Role of probiotics, prebiotics, and synbiotics in the elderly: insights into their applications. Front Microbiol 2021;12:631254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sánchez y Sánchez de la B, Martínez Carrillo BE, Aguirre Garrido JF. et al. . Emerging evidence on the use of probiotics and prebiotics to improve the gut microbiota of older adults with frailty syndrome: a narrative review. J Nutr Health Aging 2022;26:926–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bron PA, Van Baarlen P, Kleerebezem M. Emerging molecular insights into the interaction between probiotics and the host intestinal mucosa. Nat Rev Microbiol 2012;10:66–78. [DOI] [PubMed] [Google Scholar]
- 9. Sanders ME, Merenstein DJ, Reid G. et al. . Probiotics and prebiotics in intestinal health and disease: from biology to the clinic. Nat Rev Gastroenterol Hepatol 2019;16:605–16. [DOI] [PubMed] [Google Scholar]
- 10. Plaza-Diaz J, Ruiz-Ojeda FJ, Gil-Campos M. et al. . Mechanisms of action of probiotics. Adv Nutr 2019;10:S49–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Suez J, Zmora N, Segal E. et al. . The pros, cons, and many unknowns of probiotics. Nat Med 2019;25:716–29. [DOI] [PubMed] [Google Scholar]
- 12. Coutts L, Ibrahim K, Tan QY. et al. . Can probiotics, prebiotics and synbiotics improve functional outcomes for older people: a systematic review. Eur Geriatr Med 2020;11:975–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sniffen JC, McFarland LV, Evans CT. et al. . Choosing an appropriate probiotic product for your patient: an evidence-based practical guide. PLoS One 2018;13:e0209205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang C, Derrien M, Levenez F. et al. . Ecological robustness of the gut microbiota in response to ingestion of transient food-borne microbes. ISME J 2016;10:2235–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Albert MS, DeKosky ST, Dickson D. et al. . The diagnosis of mild cognitive impairment due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement 2011;7:270–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jack CR, Bennett DA, Blennow K. et al. . NIA-AA research framework: toward a biological definition of Alzheimer’s disease. Alzheimers Dement 2018;14:535–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014;30:1312–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics 2010;26:1572–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tindall BJ. The status of the name lactobacillus rogosae Holdeman and Moore 1974. Opinion 88. Judicial Commission of the International Committee on Systematics of Prokaryotes. Int J Syst Evol Microbiol 2014;64:3578–9. [DOI] [PubMed] [Google Scholar]
- 20. Zhang L, Qu H, Liu X. et al. . Comparison and selection of probiotic Lactobacillus from human intestinal tract and traditional fermented food in vitro via PCA, unsupervised clustering algorithm, and heat-map analysis. Food Sci Nutr 2022;10:4247–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ghosh TS, Arnoux J, O’Toole PW. Metagenomic analysis reveals distinct patterns of gut Lactobacillus prevalence, abundance, and geographical variation in health and disease. Gut Microbes 2020;12:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Duranti S, Ruiz L, Lugli GA. et al. . Bifidobacterium adolescentis as a key member of the human gut microbiota in the production of GABA. Sci Rep 2020;10:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tamés H, Sabater C, Royo F. et al. . Mouse intestinal microbiome modulation by oral administration of a GABA-producing Bifidobacterium adolescentis strain. Microbiol Spectr 2023;12:e02580–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Guo Y, Xie JP, Deng K. et al. . Prophylactic effects of Bifidobacterium adolescentis on anxiety and depression-like phenotypes after chronic stress: a role of the gut microbiota-inflammation axis. Front Behav Neurosci 2019;13:452616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen S, Chen L, Qi Y. et al. . Bifidobacterium adolescentis regulates catalase activity and host metabolism and improves healthspan and lifespan in multiple species. Nat Aging 2021;1:991–1001. [DOI] [PubMed] [Google Scholar]
- 26. Yamashiro K, Tanaka R, Urabe T. et al. . Gut dysbiosis is associated with metabolism and systemic inflammation in patients with ischemic stroke. PLoS One 2017;12:e0171521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Honarpisheh P, Bryan RM, McCullough LD. Aging microbiota-gut-brain axis in stroke risk and outcome. Circ Res 2022;130:1112–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. He Y, Kosciolek T, Tang J. et al. . Gut microbiome and magnetic resonance spectroscopy study of subjects at ultra-high risk for psychosis may support the membrane hypothesis. Eur Psychiatry 2018;53:37–45. [DOI] [PubMed] [Google Scholar]
- 29. Socała K, Doboszewska U, Szopa A. et al. . The role of microbiota-gut-brain axis in neuropsychiatric and neurological disorders. Pharmacol Res 2021;172:105840. [DOI] [PubMed] [Google Scholar]
- 30. Li J, Zhu S, Wang Y. et al. . Metagenomic association analysis of cognitive impairment in community-dwelling older adults. Neurobiol Dis 2023;180:106081. [DOI] [PubMed] [Google Scholar]
- 31. Wang J, Qie J, Zhu D. et al. . The landscape in the gut microbiome of long-lived families reveals new insights on longevity and aging – relevant neural and immune function. Gut Microbes 2022;14:2107288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pinto-Sanchez MI, Hall GB, Ghajar K. et al. . Probiotic Bifidobacterium longum NCC3001 reduces depression scores and alters brain activity: a pilot study in patients with irritable bowel syndrome. Gastroenterology 2017;153:448–459.e8. [DOI] [PubMed] [Google Scholar]
- 33. Chahwan B, Kwan S, Isik A. et al. . Gut feelings: a randomised, triple-blind, placebo-controlled trial of probiotics for depressive symptoms. J Affect Disord 2019;253:317–26. [DOI] [PubMed] [Google Scholar]
- 34. Veiga P, Suez J, Derrien M. et al. . Moving from probiotics to precision probiotics. Nat Microbiol 2020;5:878–80. [DOI] [PubMed] [Google Scholar]
- 35. Singh TP, Natraj BH. Next-generation probiotics: a promising approach towards designing personalized medicine. Crit Rev Microbiol 2021;47:479–98. [DOI] [PubMed] [Google Scholar]
- 36. Pan C, Ning Y, Jia Y. et al. . Transcriptome-wide association study identified candidate genes associated with gut microbiota. Gut Pathog 2021;13:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zahavi L, Lavon A, Reicher L. et al. . Bacterial SNPs in the human gut microbiome associate with host BMI. Nat Med 2023;29:2785–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The relative abundance matrix of older adults’ gut microbiome composition has been provided as supplementary materials. The raw sequencing data underlying this article cannot be shared publicly due to the privacy of individuals that participated in the study. The data will be shared on the request to the first and corresponding author.