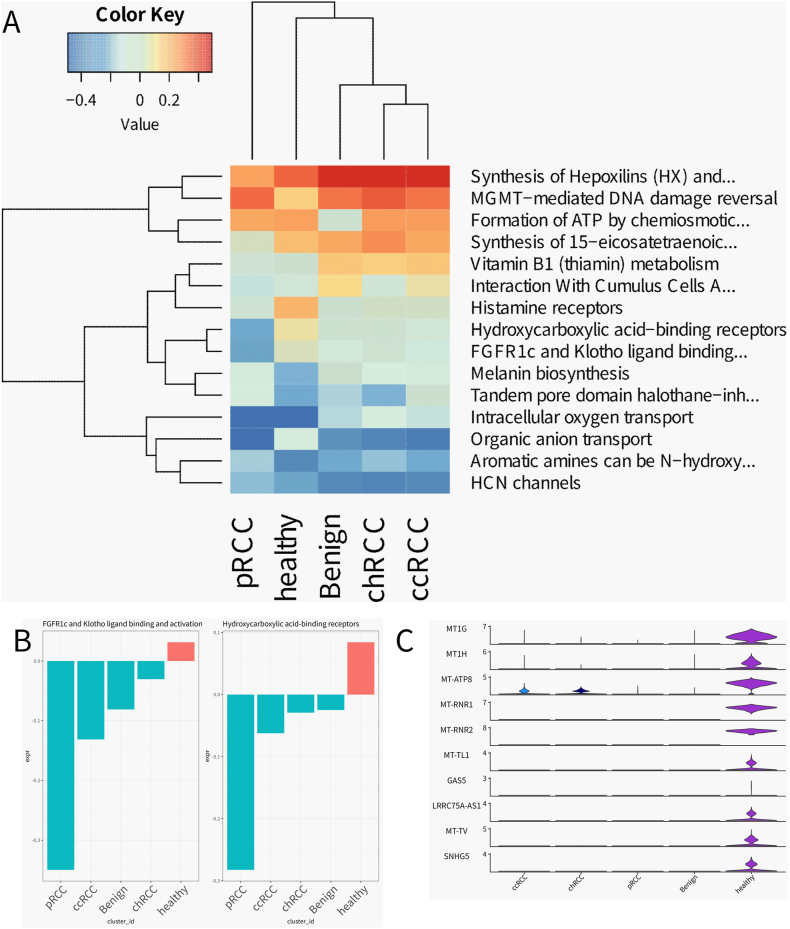
Fig. 4.
Pathway Analysis of Macrophage from all kidney sample.
A) ReactomeGSA gene set variation based pathway-level expression in the identified Macrophage. Expression values were z-score normalized by pathway.
B) Barplot generated by ReactomeGSA showing the activity of the “FGFR1c and Klotho ligand binding and activation” and “Hydroxycarboxylic acid-binding receptors” pathway in different samples. The horizontal axis shows the sample grouping, the vertical axis represents the expression value of the path, and the color represents the positive or negative expression value.
C) To compare the expression of differentially expressed genes in macrophages of healthy samples in all cell clusters.