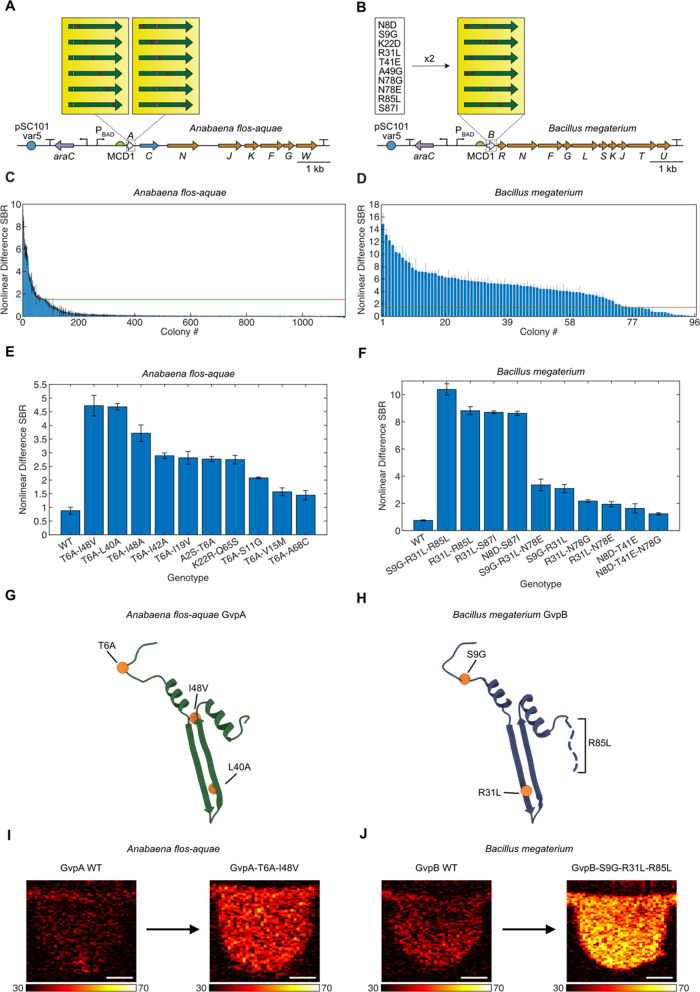
Figure 4.
Second round of directed evolution of A. flos-aquae and B. megaterium structural proteins. (A, B) Diagrams of the mutagenized A. flos-aquae and B. megaterium gene clusters used in the second round of evolution. The best two mutants of A. flos-aquae gvpA were used as parents for another scanning site saturation library, and the best ten mutants of B. megaterium gvpB (listed in figure) were used to create a paired recombination library. (C, D) Nonlinear US difference signal-to-background ratio (SBR) from all screened mutants of both clusters. Red lines indicate the difference SBR of the WT for that cluster. Error bars represent standard error. N = 3 technical replicates of one biological sample. (E, F) Nonlinear US difference SBR for the WT and top ten mutants for each cluster. Error bars represent standard error. N = 4 biological samples (each an average of 3 technical replicates). (G, H) Locations of mutations from the top mutants from (E, F) in the GvpA/GvpB structure. (PDB 8GBS and 7R1C) (I, J) Representative nonlinear US images of the brightest mutants identified in this study, as well as their respective WT parents. Scale bars 1 mm. Color bar limits in decibels (dB).