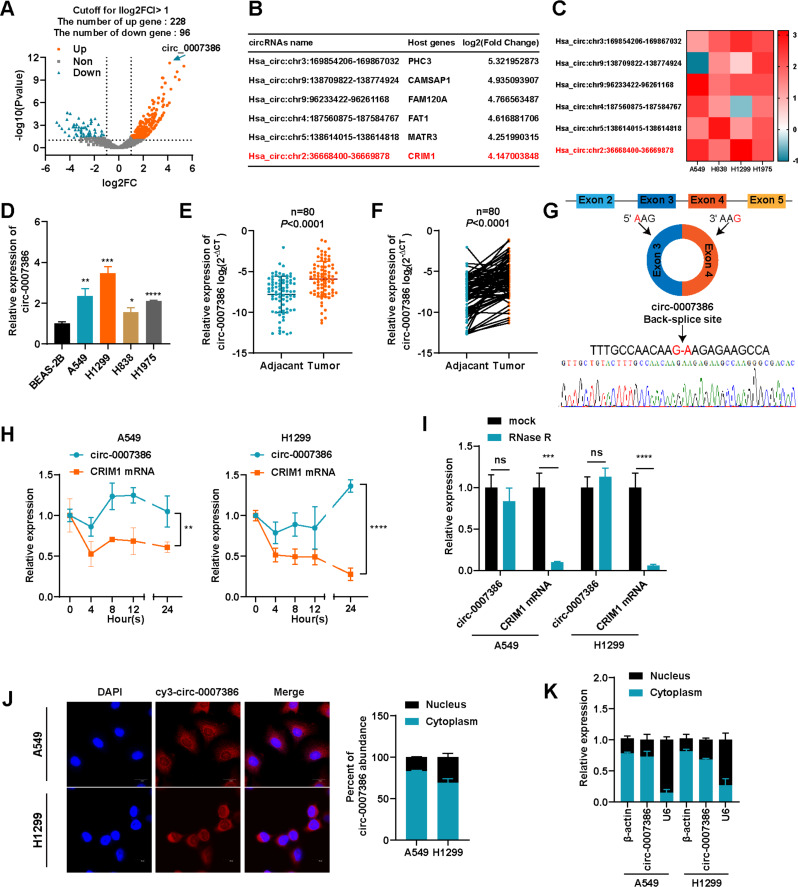
Fig. 1.
Expression and characterization of circ_0007386 in NSCLC cells and tissues. (A) Volcano plots showing the differentially expressed circRNAs in NSCLC tissue versus matched normal tissue. (B) A list of the top six dysregulated circRNAs. (C) RT-qPCR analysis was performed to compare the expression levels of the six most dysregulated circRNAs in A549, H838, H1299, and H1975 cells with their expression levels in BEAS-2B cells. (D) Relative expression levels of circ_0007386 in BEAS-2B cells and NSCLC cell lines were detected by RT-qPCR. E-F. RT-qPCR was conducted to assess the relative expression of circ_0007386 in NSCLC tissues and adjacent nontumor tissues (n = 80). G. A schematic of the genomic location and back splicing of circ_0007386 with the splicing site verified by Sanger sequencing. H. circ_0007386 and CRIM1 mRNA expression were examined by RT-qPCR in NSCLC cells after actinomycin D treatment on 0 h, 4 h, 8 h, 12 h, 24 h respectively. I. circ_0007386 and CRIM1 mRNA expression were examined in NSCLC cells after RNase R treatment. J. FISH assay showed that circ_0007386 (Cy3-labeled probe) was abundant in the cytoplasm, and DAPI was used to stain the nucleus. Images are shown at 600× magnification. Scale bar = 20 μm. K. The localization of circ_0007386 in A549 and H1299 cells detected by nuclear-cytoplasmic separation experiment. Data are shown as the means ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001