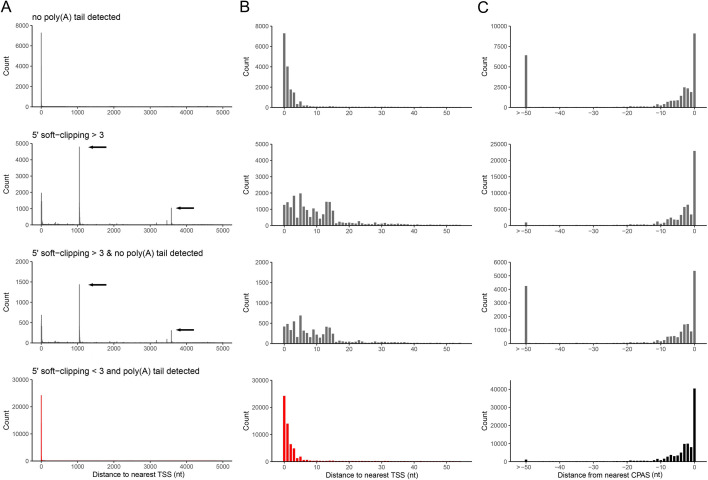
Fig 1.
Characteristics of nanopore DRS alignments. Alignments of adenovirus type 5 DRS reads were segregated according to the presence/absence of detectable poly(A) tails and the presence of soft-clipping values >3 at the 5′ end. (A–C) The genomic location and read count of (A and B) 5′ alignment ends relative to previously defined TSS or (C) 3′ alignment ends relative to previously defined CPAS were determined for each of four conditions (no poly(A) tail detected, 5′ soft-clipping >3, no poly(A) tail and 5′ soft-clipping >3, and poly(A) tail and 5′ soft-clipping ≤3) across windows of (A) 5,000 nt and (B and C) 50 nt. Black arrows indicate the location of artifact TSS derived from misalignment across splice junctions.