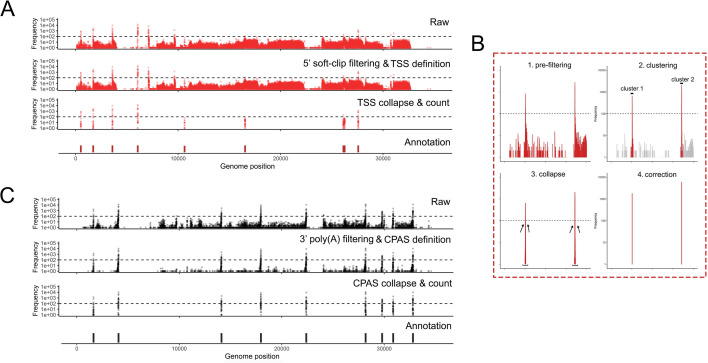
Fig 3.
Pre-filtering at 5′ and 3′ ends defines robust TSS and CPAS. NAGATA masks alignments with 5′ soft-clipping values above a user-defined value (default = 3) and, where nanopolish poly(A) output files are available, also removes alignment derived from reads without detectable poly(A) tails. (A) Visualization of TSS positions contained in the raw alignment (top), pre-filtered alignments, i.e., post-5′ soft-clipping and 3′ poly(A) tail filtering (second row), post-collapse of neighboring TSS (third row), and the existing TSS/CPAS annotation position (bottom row). (B) A specific close-up showing the pre-filtering, clustering, collapse, and correction steps. (C) Same as (A) but for CPAS.