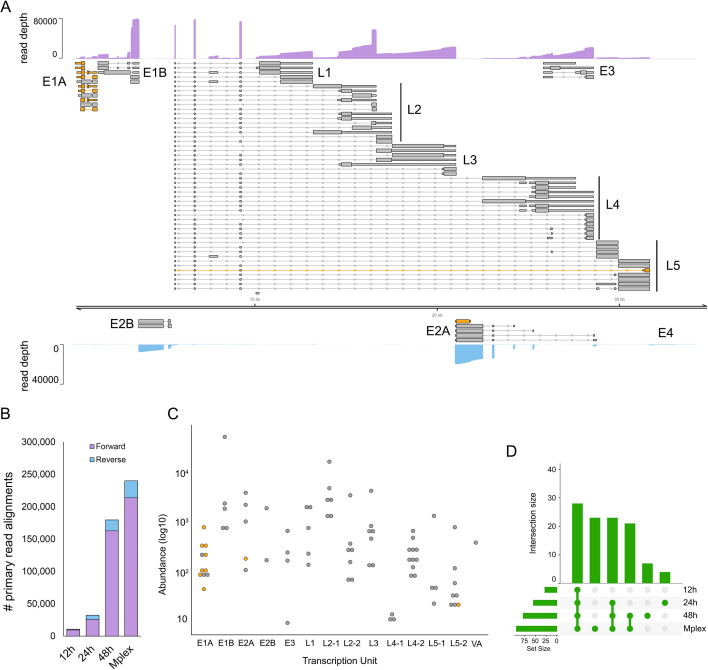
Fig 8.
Annotation of the HAdV-F41 transcriptome. (A) The reannotated HAdV-F41 transcriptome encodes 77 transcripts, of which 67 encodes 23 canonical ORFs with their UTRs defined (gray). Wide and thin boxes indicate canonical CDS domains and UTRs, respectively. A further nine transcripts (orange), putatively encoded novel or N′ terminal truncated protein isoforms. Nanopore DRS coverage plots (purple) are shown for the combined (Mplex) data set (12 + 24 + 48 hpi) in a strand-specific manner. Y-axis values indicate the read depth. (B) The number of HAdV-F41 read alignments recorded against the forward and reverse strands, separated by data set, show a strong bias toward transcription from the forward strand. (C) For each detected transcript in each transcription unit, a raw abundance count was generated using NAGATA and color-coded according to transcript classification. (D) Upset plots denoting the number of transcripts reported by NAGATA in each of the individuals, as well as the merged, data sets.