Fig 5. CRISPRi knockdown of candidate macrophage pathogenesis genes.
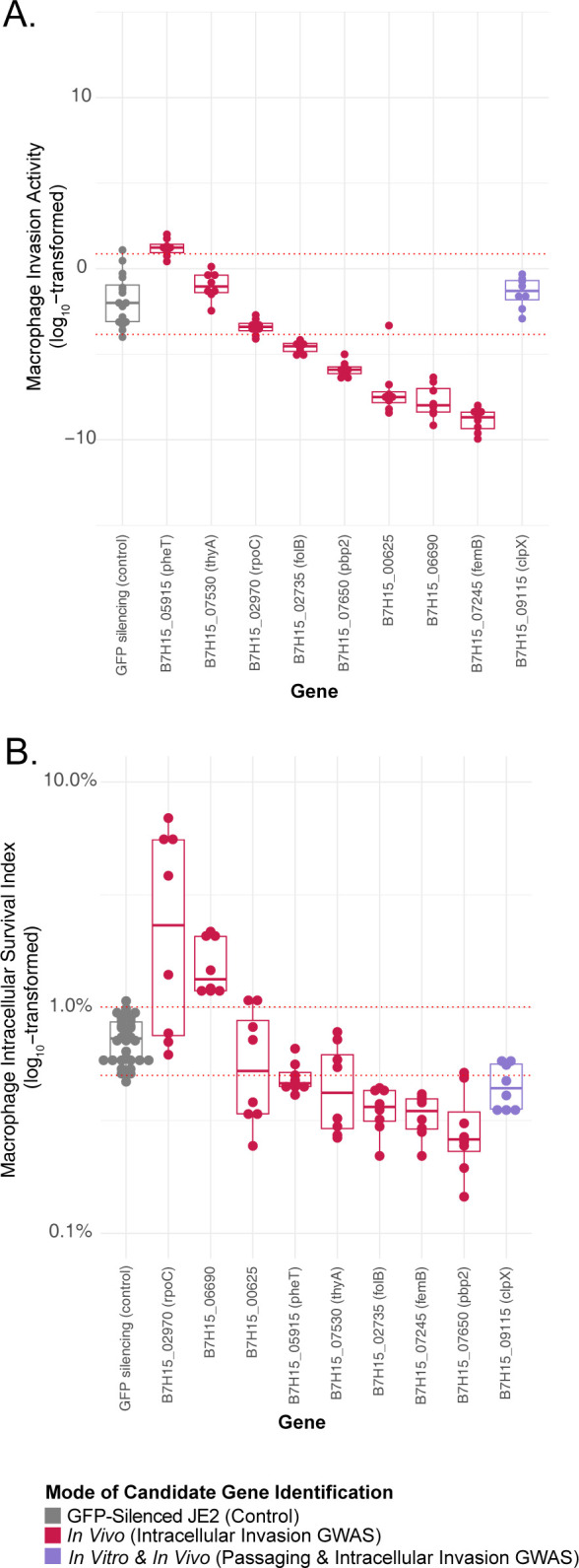
Fold-difference in measurements of macrophage invasion, normalized to in-batch testing of the parental JE2 strain carrying a GFP-targeted CRISPRi construct(A), and macrophage intracellular survival (B) for specified CRISPRi gene knockdowns. Candidate genes are grouped according to the strategy by which they were identified, and sorted within groups by median value. JE2 carrying a GFP-expressing plasmid built on the same vector backbone as silencing constructs was used as an additional control group. Boxes indicate median, 25th, and 75th percentiles with whiskers extending to the most distant value < 1.5 times the interquartile range. Red horizontal lines indicate the 95% confidence interval of all control measurements from JE2 with GFP-targeted CRISPRi, with significant mutants defined as having a median measurement outside this range. Values higher than the 95% confidence intervals indicate increased macrophage pathogenesis phenotypes, and those below the interval correspond to decreased macrophage pathogenesis phenotypes. Genome-wide association study (GWAS).
