Table 2.
Docking scores, average MM/GBSA scores, and anticancer therapeutic activity prediction values for top-scoring compounds in the ERCC1-XPF PPI binding sites 1, 2, and 3.
| ERCC1-XPF | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2D Structure | Ligand ID | Library | Anticancer activity prediction | Docking score (kcal/mol) | Post-VSW MM/GBSA average score (kcal/mol) | Short MD (10 ns) MM/GBSA average score (kcal/mol) | STDEV (10 ns) | Long MD (100 ns) MM/GBSA average score (kcal/mol) | STDEV (100 ns) | Long MD (200 ns) MM/GBSA average score (kcal/mol) | STDEV (200 ns) | |
| Site 1 |
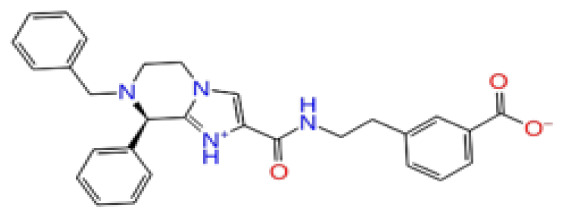
|
ZC46-0199 | CHEMDIV | 0.63 | −5.95 | −51.73 | −66.81 | 5.61 | −59.20 | 4.97 | −54.68 | 6.08 |
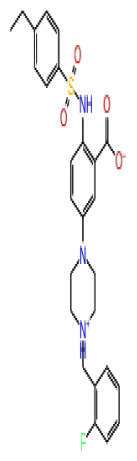
|
F684-0404 | CHEMDIV | 0.56 | −5.91 | −41.52 | −52.18 | 5.25 | −46.06 | 5.13 | −44.89 | 4.43 | |
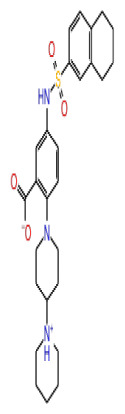
|
F294-0607 | CHEMDIV | 0.56 | −6.41 | −36.94 | −50.27 | 5.87 | −42.25 | 15.21 | −42.86 | 12.00 | |
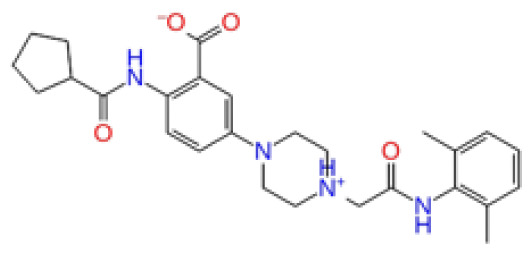
|
F687-1384 | CHEMDIV | 0.56 | −6.76 | −41.39 | −53.55 | 5.00 | −34.51 | 16.45 | −35.36 | 15.65 | |
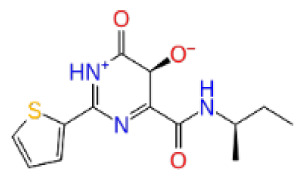
|
CHEMBL3617209 | CHEMBL | 0.83 | −4.33 | −33.70 | −31.11 | 5.05 | −33.44 | 7.20 | −33.10 | 7.47 | |
| Site 2 |
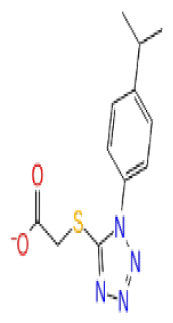
|
Z74543901 | Enamine | 0.60 | −4.29 | −22.93 | −31.76 | 5.93 | −44.54 | 6.70 | −45.89 | 5.27 |
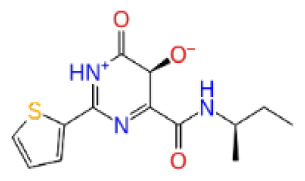
|
CHEMBL3617209 | CHEMBL | 0.83 | −3.86 | −20.02 | −22.58 | 7.57 | −39.07 | 9.63 | −39.17 | 7.44 | |
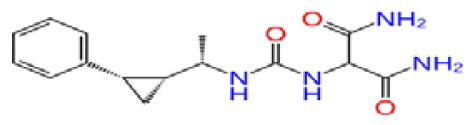
|
Z807707434 | Enamine | 0.71 | −3.85 | −38.43 | −31.06 | 7.79 | −30.71 | 5.17 | −31.28 | 9.18 | |
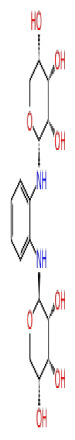
|
0527-0155 | CHEMDIV | 0.72 | −5.75 | −37.63 | −37.15 | 5.52 | −25.80 | 11.15 | −23.69 | 10.97 | |
| Site 3 |
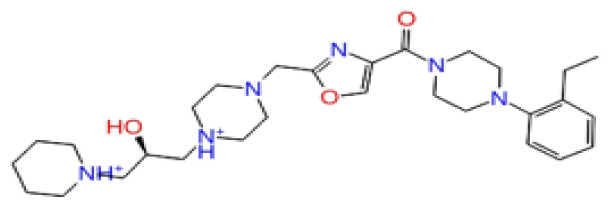
|
V008-1978 | CHEMDIV | 0.64 | −6.23 | −60.16 | −63.54 | 6.31 | −61.69 | 8.73 | −64.17 | 8.76 |
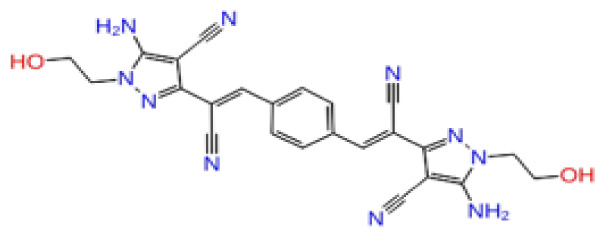
|
AG-205/36628032 | SPECS | 0.65 | −5.65 | −48.58 | −63.06 | 9.43 | −58.71 | 7.56 | −60.10 | 7.14 | |
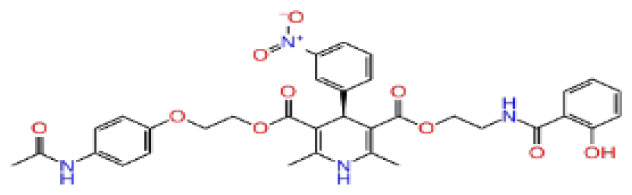
|
AQ-405/42300197 | SPECS | 0.59 | −5.59 | −58.27 | −63.27 | 8.40 | −37.39 | 10.00 | −43.40 | 12.62 | |
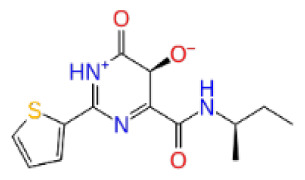
|
CHEMBL3617209 | CHEMBL | 0.83 | −3.90 | −18.75 | −30.32 | 6.69 | −41.68 | 9.19 | −43.06 | 8.02 | |
