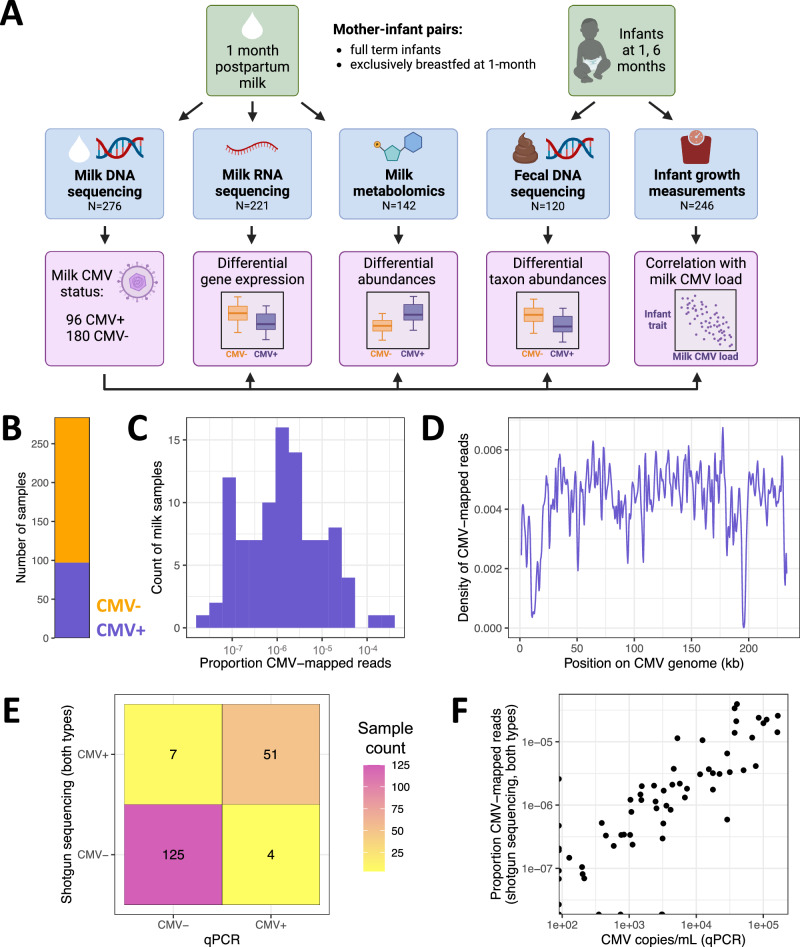
Fig. 1. Study overview and identifying CMV in human milk shotgun sequencing data.
A Study overview. B Count of milk samples identified as CMV+ (N = 97, purple) or CMV− (N = 187, orange). C The distribution of CMV-mapped DNA reads, as a proportion of all DNA reads, across milk samples that had at least one read mapped to the CMV genome. D Density of CMV-aligned reads across the CMV genome from all CMV+ milk samples. The density refers to the fraction of all CMV-mapped reads aligned to a particular region of the CMV genome. The density dips close to zero at repetitive regions in the CMV genome97. E Agreement of milk CMV status designations using shotgun DNA sequencing (y axis) vs. qPCR (x axis). F Within samples designated CMV+ by both qPCR and shotgun sequencing, the viral load estimates from the two methods were correlated (two-sided Pearson’s r = 0.88, P = 3.3 × 10−17).