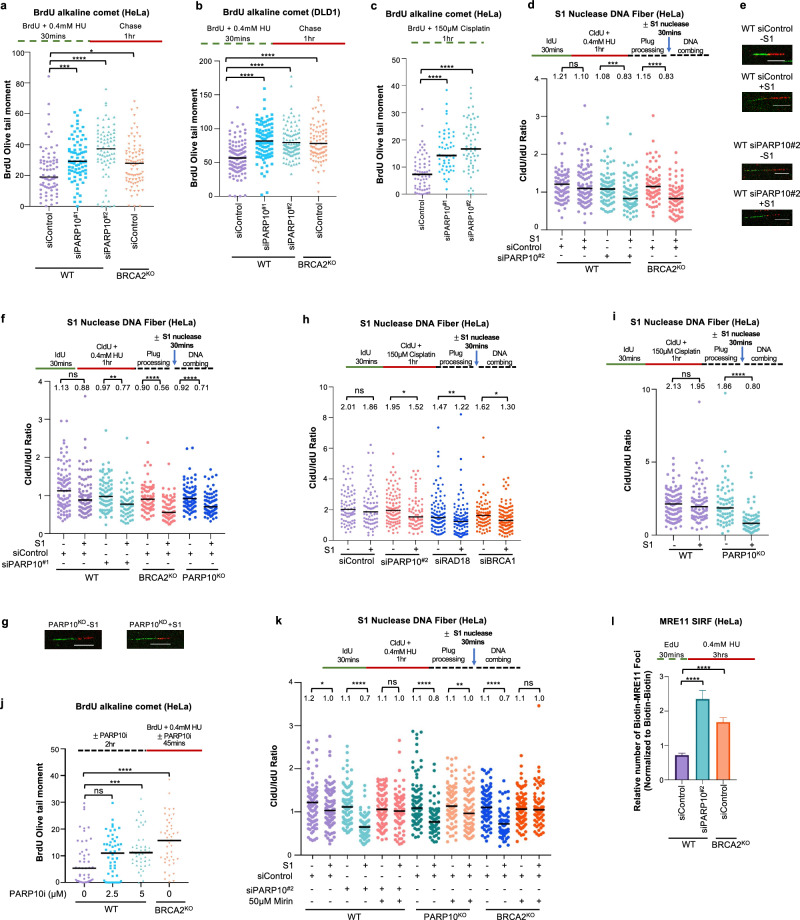
Fig. 1. Loss of PARP10 causes MRE11-mediated accumulation of ssDNA gaps.
a–c BrdU alkaline comet assays showing that PARP10 knockdown in HeLa (a, c) and DLD1 (b) cells causes accumulation of replication-associated ssDNA gaps upon treatment with 0.4 mM HU (a, b) or 150 µM cisplatin (c), similar to BRCA2 deletion. At least 50 nuclei were quantified for each condition. The median values are marked on the graph and listed at the top. Asterisks indicate statistical significance (Mann-Whitney, two-tailed). Schematic representations of the assay conditions are shown at the top. Western blots and RT-qPCR confirming PARP10 knockdown are shown in Supplementary Fig. 1a, b. d–i S1 nuclease DNA fiber combing assays showing that PARP10 knockdown (d, e, f, h) or knockout (f, g, i) in HeLa cells causes accumulation of nascent strand ssDNA gaps upon treatment with 0.4 mM HU (d–g) or 150 µM cisplatin (h, i) similar to BRCA deficiency. Quantifications (d, f, h, i) and representative micrographs, with scale bars representing 10 µm (e, g) are shown. The ratio of CldU to IdU tract lengths is presented, with the median values marked on the graphs and listed at the top. At least 63 tracts were quantified for each sample. Asterisks indicate statistical significance (Mann-Whitney, two-tailed). Schematic representations of the assay conditions are shown at the top. Western blots confirming PARP10 knockout are shown in Supplementary Fig. 1b. j BrdU alkaline comet assay showing that PARP10 inhibition using the specific inhibitor OUL35 causes accumulation of replication-associated ssDNA gaps upon treatment with 0.4 mM HU in HeLa cells. At least 44 nuclei were quantified for each condition. The median values are marked on the graph and listed at the top. Asterisks indicate statistical significance (Mann-Whitney, two-tailed). A schematic representation of the assay conditions is shown at the top. k S1 nuclease DNA fiber combing assays showing that inhibition of MRE11 endonuclease activity using the specific inhibitor mirin suppresses the accumulation of ssDNA gaps induced by treatment with 0.4 mM HU in PARP10-deficient cells. The ratio of CldU to IdU tract lengths is presented, with the median values marked on the graphs and listed at the top. At least 65 tracts were quantified for each sample. Asterisks indicate statistical significance (Mann-Whitney, two-tailed). A schematic representation of the assay conditions is shown at the top. l SIRF experiment showing that treatment with 0.4 mM HU induces binding of MRE11 to nascent DNA in PARP10-depleted HeLa cells, similar to BRCA-knockout cells. At least 92 cells were quantified for each condition. Bars indicate the mean values, error bars represent standard errors of the mean, and asterisks indicate statistical significance (t-test, two-tailed, unpaired). A schematic representation of the assay conditions is shown at the top. Source data are provided as a Source Data file.