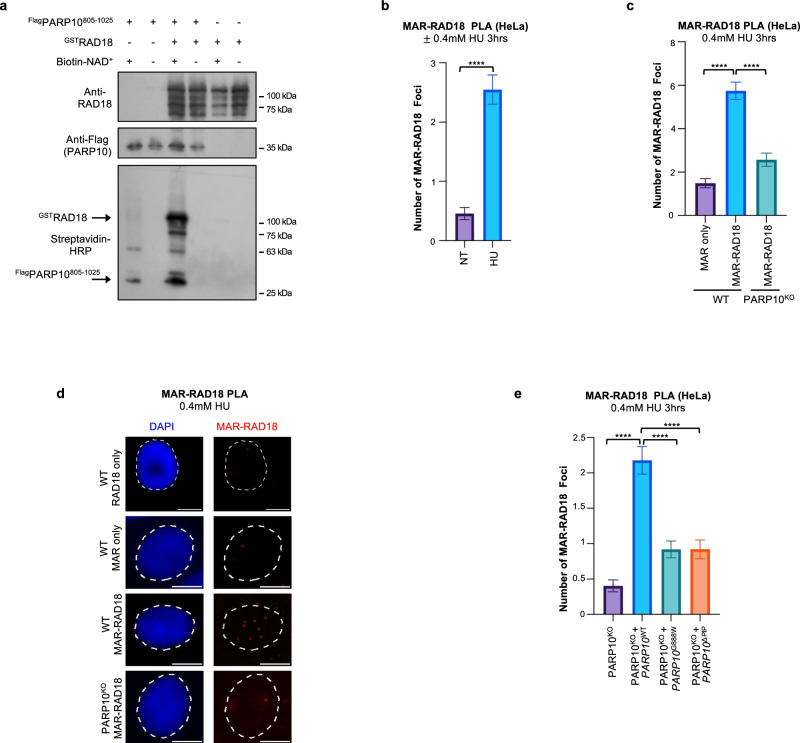
Fig. 5. Mono-ADP-ribosylation of RAD18 by PARP10.
a In vitro ADP-ribosylation enzymatic assays showing that PARP10 can MARylate RAD18. Recombinant proteins were incubated with biotin-NAD+ and substrate MARylation is detected using streptavidin-HRP blots. RAD18 and Flag (PARP10) blots are presented to show loading controls. b–d PLA assay showing that loss of PARP10 reduces the co-localization between RAD18 and mono-ADP-ribose (MAR) upon treatment with 0.4 mM HU for 3hrs in HeLa cells. Single antibody (MAR only) control is shown to confirm the specificity of the PLA signals observed. Quantifications (b, c) and representative micrographs, with scale bars representing 10 µm (d) are shown. At least 49 cells were quantified for each condition. Bars indicate the mean values, error bars represent standard errors of the mean, and asterisks indicate statistical significance (t-test, two-tailed, unpaired). e PLA assay showing the impact of PARP10 mutations on the co-localization between RAD18 and mono-ADP-ribose. Expression of wildtype PARP10, but not of the G888W catalytic site mutant or the PIP-box mutant restored the MAR-RAD18 PLA signal in PARP10KO cells. At least 60 cells were quantified for each condition. Bars indicate the mean values, error bars represent standard errors of the mean, and asterisks indicate statistical significance (t-test, two-tailed, unpaired). Source data are provided as a Source Data file.