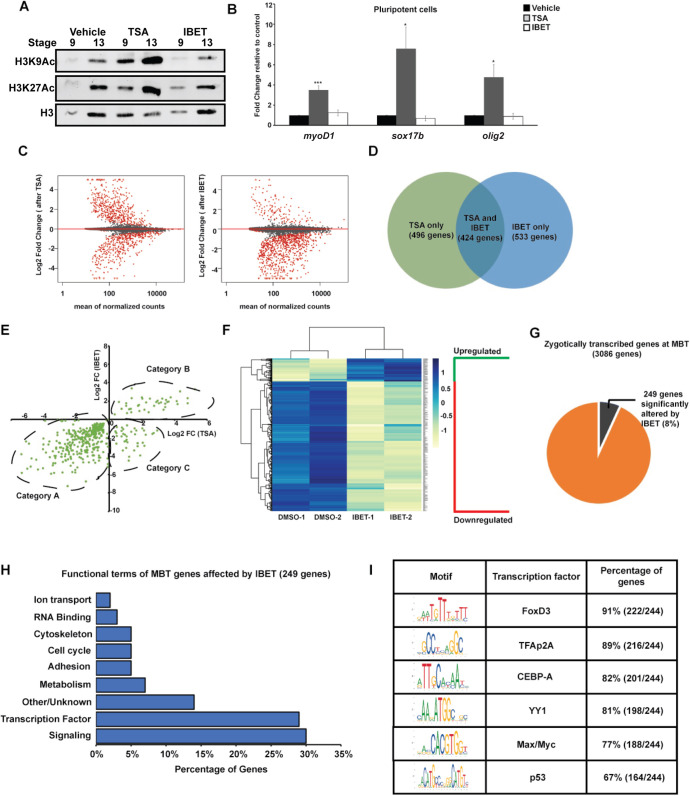
Fig. 4.
BET activity is required for activation of a subset of genes at the maternal-to-zygotic transition. (A) Western blotting for H3K9Ac and H3K27Ac with animal cap explants treated with vehicle, TSA (500 nM) or IBET (250 μM). (B) qRT-PCR for lineage markers with animal cap explants treated with vehicle, TSA (500 nM) or IBET (250 μM). *P<0.05, ***P<0.001 (unpaired, two-tailed t-test). Data are mean±s.e.m. (C) MA-plot depicting global log2 fold gene expression changes in animal cap explants after TSA (left) or IBET (right) treatment. (D) Venn diagram comparing gene sets obtained from RNA-seq of animal cap explants treated with TSA/IBET. (E) Quadrant plot comparing log2 fold changes of genes altered by TSA and IBET. Category A includes genes downregulated by both TSA and IBET, category B includes genes upregulated by both TSA and IBET and category C includes genes upregulated by TSA but downregulated by IBET. (F) Heatmap of genes changed only by IBET but not TSA. (G) Pie chart depicting the genes zygotically transcribed at MZT that are affected by IBET. (H) Functional terms for zygotically transcribed mid-blastula transition (MBT) genes affected by IBET show enrichment for signaling molecules and transcription factors. (I) Motif analysis of promoters of genes altered by IBET at MBT. Enrichment is observed for transcription factor binding of known factors that have been previously characterized to be involved with BET proteins and have known functions in stem cell maintenance and neural crest formation.