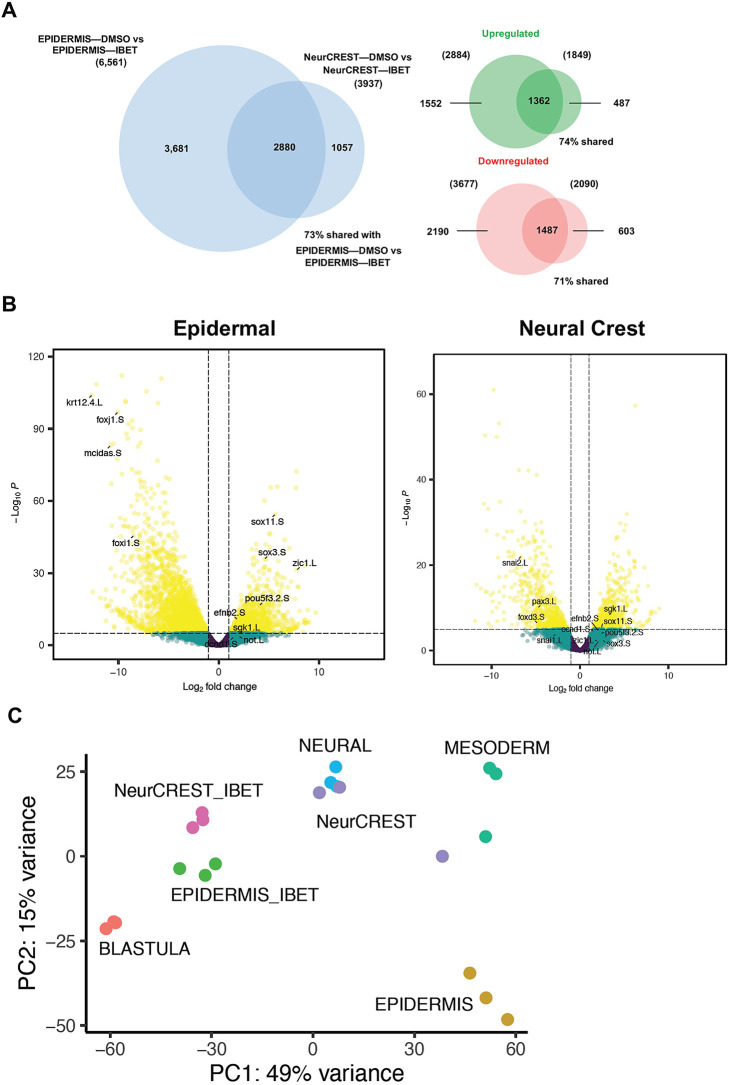
Fig. 6.
Transcriptome changes in response to BET inhibition are shared across state transitions. (A) Venn diagrams depicting the overlap between genes found to be significantly altered using DESeq2 when comparing stage 13 epidermal explant response to IBET (250 μM) with stage 13 neural crest-reprogrammed explant response to IBET. Larger diagram compares total genes sets; smaller diagrams compare upregulated or downregulated gene sets. (B) Volcano plots of genes differentially expressed in stage 13 epidermal and neural crest explants in response to IBET. Genes noted are those that are downregulated in epidermal cells only (krt12.4.L, foxj1.S, mcidas.S, foxi1.S), downregulated in neural crest only (snai2.L, snai1.L, foxd3.S, pax3.L), and genes which are upregulated in both sets (efnb2.S, ccnd1.S, sgk1.L, sox11.S, sox3.S, zic1.L, pou5f3.2.S, not.L). (C) PCA plot comparing the variance between the transcriptomes of IBET-treated explants (EPIDERMIS_IBET, NeurCREST_IBET) and that of blastula and several other lineages (Neural, Mesoderm, Epidermis, NeurCREST). Aside from blastula-stage explants, explants of all other lineages were collected at stage 13.