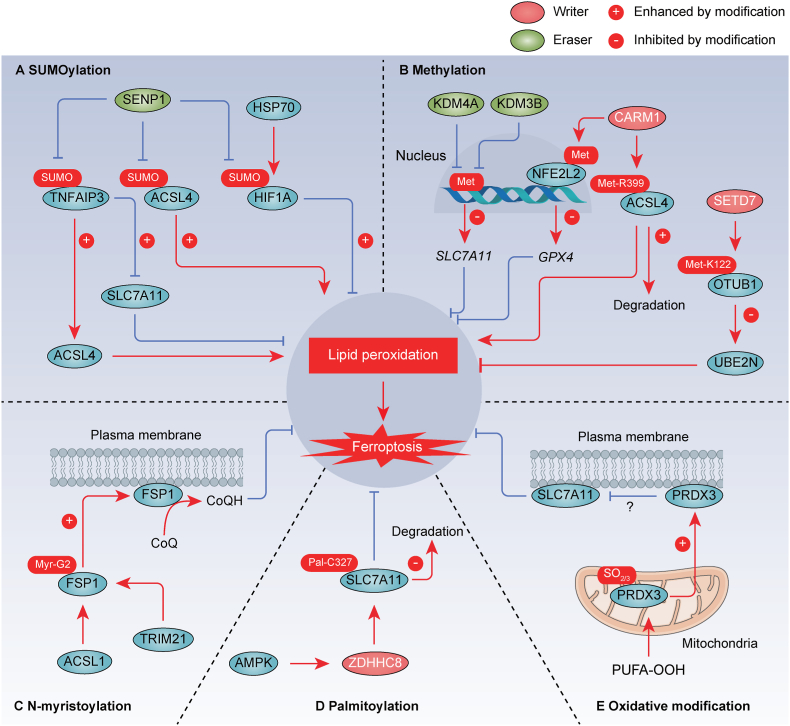
Fig. 5.
SUMOylation, methylation, N-myristoylation, palmitoylation, and oxidative modification in ferroptosis. A. SUMOylation modifies various regulatory factors (e.g., TNFAIP3/A20, ACSL4, and HIF1A) of ferroptosis. B. Methylation serves as a modifying mechanism for a range of proteins, including histone, NFE2L2/NRF2, ACSL4, and OTUB1, influencing the processes of ferroptosis. C. N-myristoylation of FSP1 at Gly2 recruits it to the plasma membrane, where it functions as an oxidoreductase to generate CoQH2 parallel to GSH system. D. ZDHHC8 catalyzes the S-palmitoylation of SLC7A11 specifically at C327 and diminishes the ubiquitination levels to stabilize SLC7A11. E. PRDX3 yields cysteine oxidation to sulfinic acid (-SO2) and sulfonic acid (-SO3), which induces its translocation from mitochondria to plasma membrane, where it inhibits cystine uptake. The promotion or inhibition of protein function by the modification is indicated by a positive (+) or negative (−) sign, respectively. SUMO, SUMOylation. Met, methylation. Myr, N-myristoylation. Pal, palmitoylation.