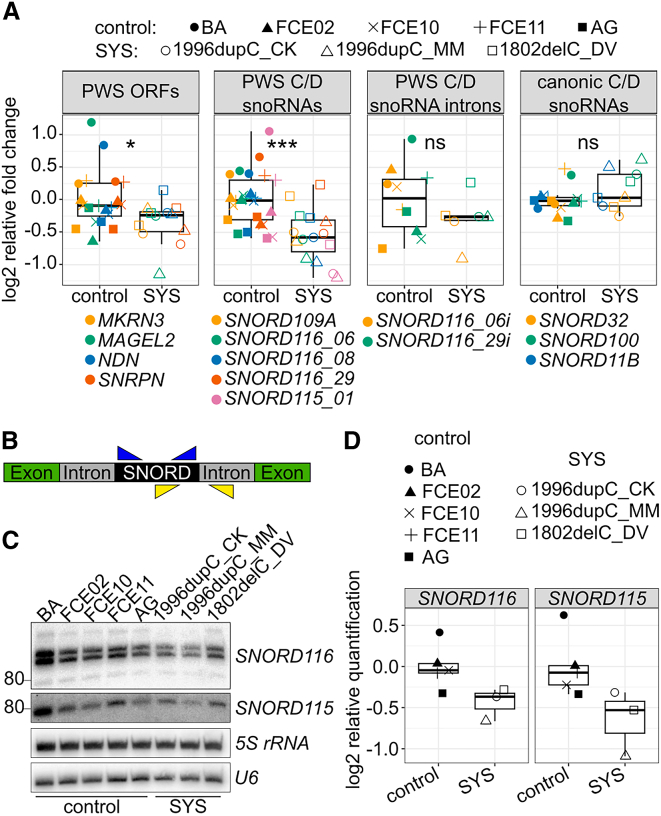
Figure 6.
Influence of MAGEL2 frameshift mutations on gene expression, assessed in SYS smNPCs compared to healthy controls
(A) RT-qPCR analysis of genes of interest (GOI) within and outside chromosome 15q11–q13. Relative expression was calculated with primer efficiency normalized N0 values (linRegPCR50). N0 values were normalized to the geometric mean of housekeeping genes TBP, GAPDH, and HPRT and to the geometric mean of healthy control samples. Genes were combined into groups depending on type and genomic localization. Wilcoxon tests were applied employing rstatix.
(B) Scheme of primer position to detect snoRNAs before (yellow) and after (blue) splicing and processing.
(C) Northern blot analysis of 19 and 30 copies of SNORD116 and SNORD115, respectively.
(D) Northern blot bands were quantified employing Fiji and normalized to geometric mean areas of 5S rRNA and U6 snRNA signals.