Abstract
Vestimentiferan tubeworms that thrive in deep-sea chemosynthetic ecosystems rely on a single species of sulfide-oxidizing gammaproteobacterial endosymbionts housed in a specialized symbiotic organ called trophosome as their primary carbon source. While this simple symbiosis is remarkably productive, the host-symbiont molecular interactions remain unelucidated. Here, we applied an approach for deep-sea in situ single-cell fixation in a cold-seep tubeworm, Paraescarpia echinospica. Single-cell RNA sequencing analysis and further molecular characterizations of both the trophosome and endosymbiont indicate that the tubeworm maintains two distinct metabolic “microniches” in the trophosome by controlling the availability of chemosynthetic gases and metabolites, resulting in oxygenated and hypoxic conditions. The endosymbionts in the oxygenated niche actively conduct autotrophic carbon fixation and are digested for nutrients, while those in the hypoxic niche conduct anaerobic denitrification, which helps the host remove ammonia waste. Our study provides insights into the molecular interactions between animals and their symbiotic microbes.
Deep-sea tubeworms harbor two “microniches” for their endosymbionts, enabling efficient carbon fixation and waste removal.
INTRODUCTION
The vestimentiferan tubeworms (Siboglinidae and Polychaeta) that live in deep-sea chemosynthetic ecosystems, such as hydrothermal vents, cold seeps, and whale falls (1–4), often lack a digestive system and have no mouth, digestive tract, nor anus (1). Instead, they rely on sulfide-oxidizing gammaproteobacterial endosymbionts to meet their metabolic needs (5–7). The endosymbionts are housed in a specialized organ called the trophosome (8), which consists of numerous highly vascularized lobules ranging from 100 to 300 μm in diameter (9), akin to the vertebrate liver lobule. The trophosome locates deep within the trunk of the tubeworm and has no direct contact with the ambient environment (8). Therefore, the tubeworm must transport oxygen, hydrogen sulfide, and inorganic carbon compound in large quantities to the “remotely located” trophosome where the numerous symbionts reside and use these raw ingredients to produce organic matter (10–12); it must also remove the waste products from the trophosome (13, 14). To do so, first, they extract oxygen, carbon dioxide, and sulfide from their surrounding environment through their plume or roots (15, 16). These molecules are transported in the coelomic fluid and to the trophosome lobules where the endosymbionts reside. It has been hypothesized that there are molecular gradients in the trophosome lobule (17, 18) to facilitate its various metabolic functions such as nutrient production, host-symbiont metabolites exchange, and elimination of waste products (9, 19).
Despite the complexities of this arrangement, the tubeworm-microbes symbiosis is remarkably adaptive and productive (20, 21). Tubeworms often form high-density communities and dominate deep-sea chemosynthetic ecosystems (22, 23). Some species, such as the giant tubeworm Riftia pachyptila, can grow extraordinarily fast, with the highest recorded annual growth of more than 85 cm in tube length (20). This enormous productivity indicates an effective metabolic process underpinned by many symbiosis-specific adaptations of both the symbiotic microbes and the host.
The endosymbiont’s metabolism has been extensively studied, mainly using metagenomic and metaproteomic approaches (18, 24–27). Each tubeworm species often associates with a single 16S ribosomal RNA (rRNA) phylotype of endosymbiotic bacteria (28), but the symbionts exhibit remarkable metabolic versatility and redundancy (18, 27). Symbionts from the same host animal express enzymes of both aerobic (Calvin-Benson cycle) and anaerobic [reductive tricarboxylic acid (rTCA) cycle] carbon fixation pathways (18, 27). Moreover, the proteins involved in using hydrogen sulfide and thiosulfate as electron donors and proteins using nitrate and oxygen as electron acceptors were simultaneously expressed by the same symbiont population (29). Furthermore, individual symbiont cells exhibit pronounced morphological diversity, which may represent metabolically dissimilar stages of a physiological differentiation process (9, 18).
From the collective evidence, specific molecular interactions in the tubeworm host play a crucial role in regulating the metabolism and differentiation of the endosymbionts. For instance, the host’s trophosome produces genes that generate proteins for transporting the substrates necessary for the symbiont’s chemosynthesis. These include specialized hemoglobin that can bind both sulfide and oxygen (30, 31) as well as carbonic anhydrase for CO2 uptake (12, 32, 33). The host also produces various proteins associated with host-symbiont interaction, symbiont digestion, and nutrient harvests, such as immune receptors (34, 35), protein lyases (36–38), and glycan lyases (39). Nevertheless, these studies have not been able to measure the relative abundance of different cell types from the absolute expression per cell. Hence, the details of these host-symbiont interactions still need to be elucidated.
Understanding the molecular-level interactions between hosts and their symbionts depends on comprehending the gene expression patterns of each cell type that constitutes the trophosome. Furthermore, it is essential to gain integrated knowledge of the gene expression profiles of both the host and symbiont. In the present study, we used single-cell RNA sequencing (scRNA-seq) to investigate how host-symbiont interactions are regulated in Paraescarpia echinospica (Fig. 1, A and B), which is the vestimentiferan tubeworm commonly found in west Pacific deep-sea cold seeps (27, 38). We developed a deep-sea animal fixation apparatus and modified the acetic-methanol (ACME) single-cell fixation solution (40), which allowed us to collect and fix samples of tubeworms in situ for downstream single-cell sequencing analysis. We then conducted scRNA-seq using the 10X Chromium platform to analyze the gene expression profiles of the major cell types in P. echinospica’s trophosome. The scRNA-seq data, combined with a detailed molecular characterization of key genes and metabolites from both the host and symbionts, provide insights into the interactions between the tubeworm and its intracellular symbiont, thus deepening our understanding of such symbiotic interactions and the environmental adaptation of a holobiont.
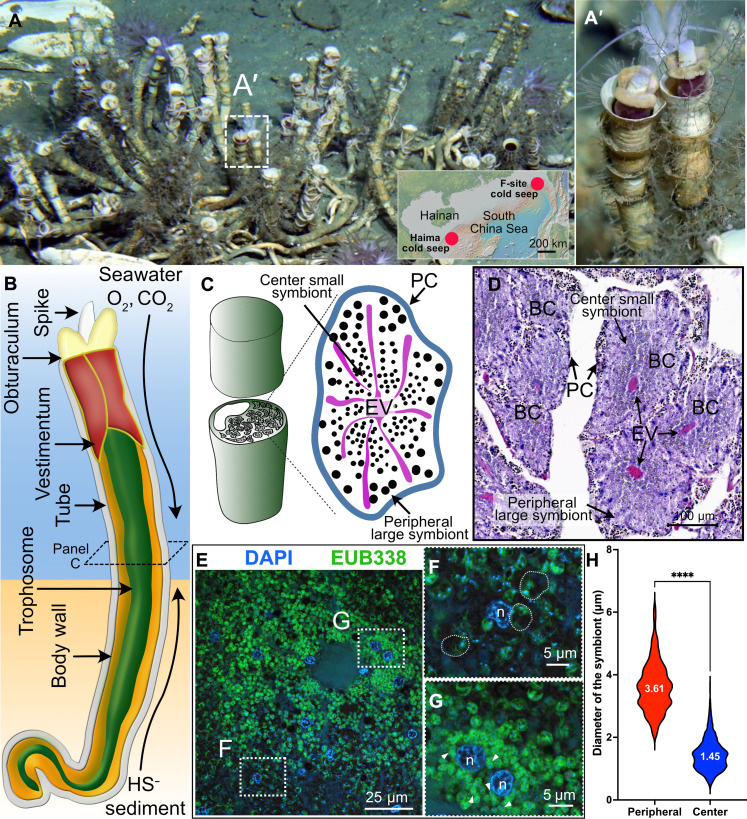
Fig. 1. Natural habitat, gas exchange system, trophosome, and endosymbionts of the deep-sea siboglinid tubeworm P. echinospica.
(A) Photograph of a colony of P. echinospica individuals in the field. Each tube is roughly 30 cm in length. The photo was taken at the Haima cold seep field by the Haima ROV crew. The inserted map shows the regional location map of the two research sites. (B) Diagram of the gas exchange system of P. echinospica. The tubeworm absorbs oxygen and carbon dioxide from the ambient seawater through its plume. It also sucks in sulfide through its posterior region from the sediment. (C) Diagram showing the cross section of each trophosome lobule, with large symbionts located at the lobule periphery and small symbionts at the lobule center. (D) H&E staining of a cross-section of the trophosome of an adult P. echinospia tubeworm. (E) FISH analysis of the symbionts. (F and G) Amplified regions of the periphery and center of a trophosome lobule, demonstrating the morphological variation of the symbionts. The semitransparent dashed lines in (F) outline three typical periphery large symbionts. (H) Statistical analysis of the size variation of bacteriocytes located in the periphery bacteriocytes and center of a lobule (P < 0.001, t test, n-periphery = 449, n-center = 1304).
RESULTS AND DISCUSSION
The trophosome of tubeworm P. echinospica
Similar to R. pachyptila and Lamellibrachia luymesi, the P. echinospica trophosome is a tissue with numerous interconnecting lobules in the trunk coelom, as shown by hematoxylin and eosin (H&E) staining (fig. S1A and Fig. 1C). The diameter of each lobule ranges from ~100 to 300 μm. Each lobule is enclosed by a single-layer peritoneal cell membrane and transversed by an efferent vessel system (Fig. 1C). Bacteriocytes are the symbiont-containing cells that reside in the lobules of the trophosome, and the symbionts belong to a single 16S rRNA phylotype (27) of sulfur-oxidizing Gammaproteobacteria (fig. S1B). The density of endosymbionts varies within each trophosome lobule, as indicated by a stronger hematoxylin staining in the center of a lobule compared to its periphery (Fig. 1D). Analyses using 16S fluorescence in situ hybridization (FISH) show morphologically distinct subtypes of symbionts that seem to preferentially appear in different lobular regions (Fig. 1, E to H): Larger cocci-shaped symbionts tend to appear in the periphery (Fig. 1F; average diameter of ~3.61 μm), while small rod-shaped symbionts are found near the center of each lobule (Fig. 1G; average diameter of ~1.45 μm). Similar trends of different symbiont subpopulations preferentially residing in different regions of the trophosome have been reported in other vestimentiferan tubeworms, such as R. pachyptila; it has been suggested that R. pachyptila’s symbionts also comprise subpopulations with distinct metabolic profiles and cellular functionalities (18, 41). Given this evidence, the trophosome tissue likely interacts with symbionts in a cell type–specific manner to maintain and regulate different subpopulations, but the molecular details of such interactions were largely unknown.
Deep-sea in situ fixation and scRNA-seq of the trophosome of P. echinospica
To faithfully capture the single-cell transcriptional information and investigate the host-symbiont interaction in P. echinospica, we developed a deep-sea in situ scRNA-seq workflow (Fig. 2A). On the basis of the ACME protocol reported by García-Castro et al. (40), we developed and tested a high-density ACME (HD-ACME) single-cell fixation solution, which is compatible with our deep-sea in situ animal fixation apparatus, also developed in-house (fig. S2, A and B). During the R/V Kexue 2020 South China Sea Cold-Seep expedition, we successfully conducted in situ P. echinospica sample collection and fixation at the “F-site” cold seep (22.11618°N, 119.2853°E; depth of 1145 m) with the deep-sea remotely operated vehicle (ROV) Faxian (fig. S2C). We then sorted individual intact cells from a single tubeworm trophosome specimen using fluorescence-activated cell sorting (FACS) in the laboratory (fig. S3A and Fig. 2B). Subsequently, we created scRNA-seq libraries with the 10X Chromium platform. Further experimental details can be found in Materials and Methods.
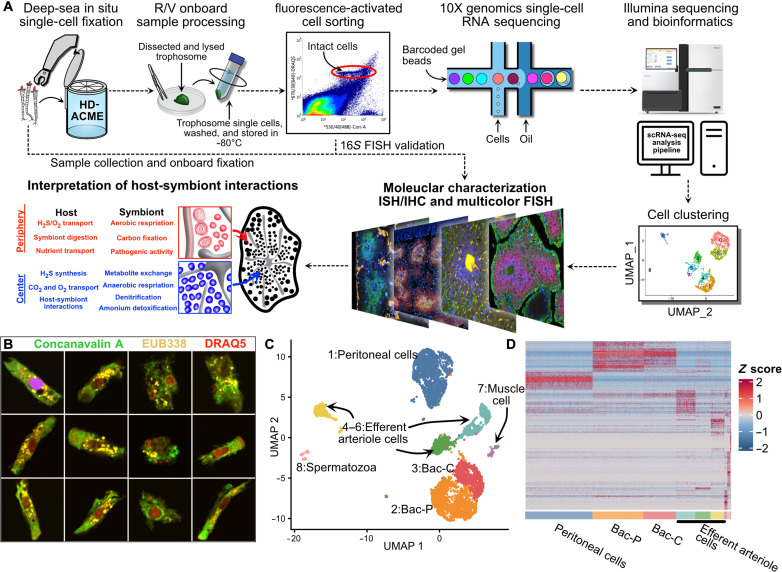
Fig. 2. Deep-sea in situ single-cell analysis of P. echinospica trophosome.
(A) Diagram showing the overall workflow of the present study. (B) FISH analysis of FACS-isolated bacteriocytes. (C) Uniform manifold approximation and projection (UMAP) of the major trophosome cell clusters. (D) Heatmap showing the expression pattern of cell cluster markers.
Eight cell clusters (fig. S4), along with their corresponding marker genes, are identified and characterized (Fig. 2, C and D). Those clusters include peritoneal cells, efferent arteriole (EA) cells, and two types of bacteriocytes constituting the trophosome. We also find muscle and spermatozoa cell clusters based on their markers (table S1) and expression patterns (fig. S5); because these two cell types are likely not involved in symbiosis and outside the scope of the present study, we left their analysis to a future endeavor.
The major nonsymbiotic cell types in the trophosome of P. echinospia
In the trophosome, we found four nonsymbiotic cell populations distinguishable by their gene markers. These gene markers offer clues as to the potential functions of the respective cell populations. Cluster 1 contains peritoneal cells found in the peritoneum of the trophosome (Fig. 3, A to G), marked by expression of the gene encoding collagen triple helix repeat containing protein 1 (table S2) and laminin subunit alpha (fig. S6A). The peritoneal cells explicitly expressed genes that encode components of extracellular hemoglobin (Fig. 3, D and E, fig. S6B, and table S2), a protein complex that can simultaneously and reversibly bind oxygen and hydrogen sulfide (10); this vital mechanism responsible for transporting these essential small molecules in the tubeworm’s tissue is a prime example of this animal’s adaptation to the deep-sea chemosynthetic ecosystem. In R. pachyptila, the extracellular hemoglobin is assembled from four globin subunits (A1, A2, B1, and B2), among which A2 and B2 subunits bind to sulfide via their free cysteine residues (42). The P. echinospia genome also has all four globin subunits (38), and it encodes a single copy of HbA2 and HbB2 but 26 copies of HbB1 (38), of which 9 of these HbB1s have free cysteine residues and may have sulfide-binding capability (38). Our scRNA-seq and ISH data showed that transcripts encoding oxygen-binding and sulfide-binding subunits were highly expressed in the peritoneal cells (Fig. 3B, fig. S6B, and table S2). Furthermore, most (15 of 16) P. echinospia extracellular hemoglobin components contained signal peptides without transmembrane domains (fig. S7), suggesting that these proteins may also be secreted in the coelom fluid. The expansion and elevated expression of sulfide-binding globin subunits may be related to P. echinospica’s adaptation to methane seeps because the worm must take sulfide from the sediment. These results also highlight the critical role of the trophosome peritoneum in regulating chemosynthetic gas transportation into the trophosome lobules for the symbionts’ use.
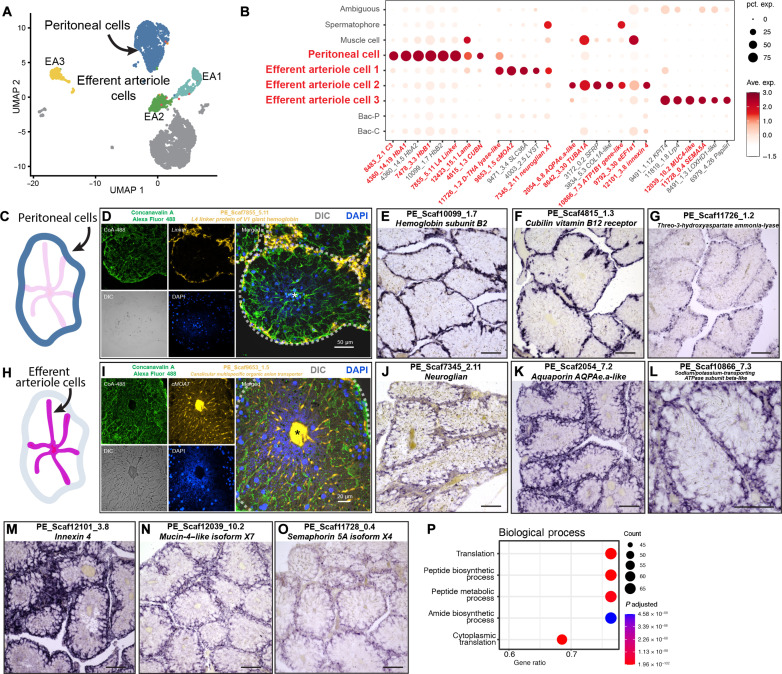
Fig. 3. Characterization of peritoneal cells and EA cells of P. echinospica trophosome.
(A) UMAP representation of peritoneal cell cluster and EA cell clusters. (B) Expression profiles of the cell markers that are specific or enriched in the peritoneal cell and EA cell populations. The sizes of the circles represent the percentages of cells in those clusters that expressed a specific gene. Genes shown in red were examined by ISHs. (C to G) FISH and colorimetric ISH of peritoneal cell population markers. DIC, differential interference contrast. (H to O) Colorimetric ISH of EA cell population markers. (P) Enriched GO biological process terms in the EA3 cell populations. The scale bars in the colorimetric ISH analyses represent 100 μm.
We identified three cell clusters representing the trophosome EA (Fig. 3H) based on cluster markers’ functionality and expression patterns (Fig. 3, B and H to O, and fig. S8). These cells form each trophosome lobule’s central-axial efferent blood vessel system and facilitate blood flow from the periphery to the center (9, 43). In cells EA1 and EA2, genes that encode transporter proteins for nutrients and waste, such as canalicular multispecific organic anion transporter 2 (PE_Scaf9653_1.5, Fig. 3I), aquaporin (PE_2054_6.8, Fig. 3K), and subunit of sodium-potassium–transporting adenosine triphosphatase (ATPase) (PE_Scaf10866_7.3, Fig. 3L), were up-regulated. In addition, the genes that encode regulatory proteins (neuroglian, PE_Scaf7345_2.11, Fig. 3J) were up-regulated in EA1 cells; the genes that encode structural proteins, such as α-tubulin (PE_Scaf8642_3.30, fig. S7A) and innexin 4 (PE_Scaf12101_3.8, Fig. 3M), were up-regulated in EA2 cells. These results indicate that EA1 and EA2 cells are distinct cell types, structurally akin to endothelial cells that comprise the walls of efferent blood vessels of the human liver (44).
The EA3 cells expressed genes that encode various proteins, such as the extracellular matrix protein mucin-4–like protein (PE_Scaf12039_10.2, Fig. 3N) and regulatory proteins like low-density lipoprotein receptor–related protein 4 (PE_Scaf11819_1.8, Fig. 3B) and semaphorin 5A (PE_Scaf11728_0.4, Fig. 3O). Furthermore, these cells highly expressed genes that encode ribosomal protein subunits, indicating that peptide and protein synthesis were highly active in this cell type (Fig. 3P). In a previous study (45), a similar cell type called the interlaminar cell was observed in the symbiotic tissue of the deep-sea mussel Gigantidas platifrons. These cells were found in the tissue where bacteriocytes were attached between two sheets of gill laminae. The interlaminar cells of G. platifrons also highly expressed genes that encode ribosomal proteins, indicating elevated metabolic and protein synthesis activities. These findings suggest that localized nutrient processing and protein synthesis within the trophosome tissue could be a convergent adaptive mechanism among deep-sea animals associated with endosymbionts.
Bacteriocytes and “microniches” in the trophosome
Our scRNA-seq data revealed two clusters of bacteriocytes in the P. echinospica trophosome with differentiating gene expression patterns [clusters bacteriocyte-periphery (abbreviated as Bac-P) and bacteriocyte-center (abbreviated as Bac-C); Fig. 4, A and B], indicating each subpopulation’s specific function within the trophosome. Note that the method we used for scRNA-seq was a polyadenylate capture approach that mostly captures transcripts from the host bacteriocyte cells, not the symbionts.
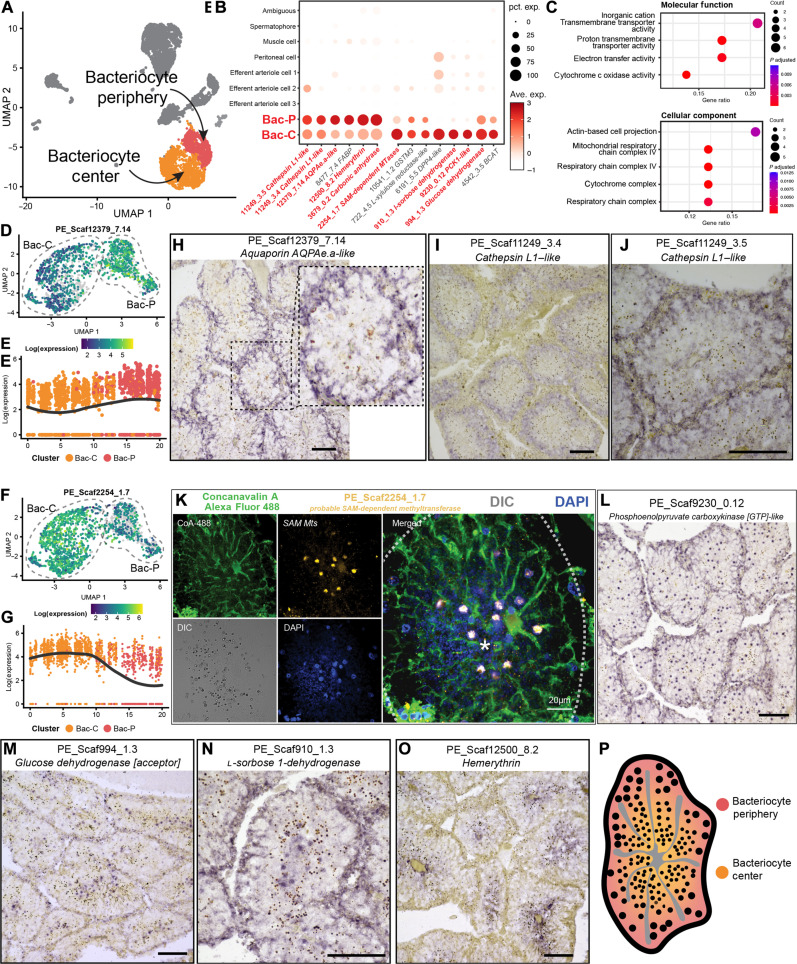
Fig. 4. Characterization of bacteriocyte cells of P. echinospica trophosome.
(A) UMAP representation of the two bacteriocyte populations: the Bac-P and the Bac-C. (B) Expression profiles of the cell markers that are specific or enriched in two bacteriocyte populations. The sizes of the circles represent the percentages of cells in those populations that expressed a specific gene. Genes shown in red were examined by ISH. (C) Enriched GO molecular function and cellular component terms in the Bac-C populations. (D and E) Gene expression trajectory analysis of Bac-P marker aquaporin AQPAe.a-like (PE_Scaf12379_7.14), showing a gradient pattern of transitioning from higher Bac-P to lower Bac-C cells. (F and G) Gene expression trajectory analysis of Bac-C marker probable SAM Mt (PE_Scaf2254_1.7), showing a gradient pattern of transitioning from higher Bac-C to lower Bac-P cells. (H to J) Colorimetric ISH analyses of Bac-P population markers, showing the genes encoding AQPAe.a-like (PE_Scaf12379_7.14), cathepsin L1–like (PE_Scaf11249_3.4), and cathepsin L1–like (PE_Scaf11249_3.5) are all highly expressed in the Bac-P cells. (K) FISH analysis of Bac-C gene marker probable SAM Mt (PE_Scaf2254_1.7), showing that the gene is expressed in the nucleus of Bac-C cells. The semitransparent dashed line marks the outline of the trophosome lobule, and the asterisk indicates the center of the lobule. (L to N) Colorimetric ISH analyses of Bac-C markers, showing that the genes encoding phosphoenolpyruvate carboxykinase [GTP]-like (PE_Scaf9230_0.12), glucose dehydrogenase [acceptor] (PE_Scaf994_1.3), and l-sorbose 1-dehydrogenase (PE_Scaf910_1.3) are highly expressed in the Bac-C cells. (O) Colorimetric ISH analysis of the gene encoding hemerythrin (PE_Scaf12500_8.2), showing that the gene is highly expressed in the Bac-C cells. (P) Diagram showing the proposed spatial distribution of two bacteriocyte populations in a single trophosome lobule. The scale bars in the colorimetric ISH analyses represent 100 μm.
Bac-P cells expressed relatively higher levels of lysosomal cysteine proteases cathepsins (PE_Scaf11249_3.4 and PE_Scaf11249_5.8, Fig. 4B) transcripts. Cathepsins are evolutionarily conserved molecular tools used by various animal hosts to digest their symbiotic microbes (36, 38, 46). Bac-P cells also expressed higher levels of genes encoding subunits of cytochrome c oxidase (table S3), the critical enzyme in the aerobic respiratory electron transport chain (47). Additional gene ontology (GO) analysis showed that GO terms related to cellular aerobic respiration were enriched in Bac-P (Fig. 4C).
In a single-cell trajectory analysis, we found the gene expression abundance of Bac-P marker aquaporin AQPAe.a-like (PE_Scaf12379_7.14) gene showed a gradient pattern transitioning from higher Bac-P to lower Bac-C cells (Fig. 4, D and E). Analysis of the Bac-C marker S-adenosylmethionine (SAM)–dependent methyltransferase (SAM Mt) (PE_Scaf2254_1.7) gene expression trajectory also showed a similar pattern of gradated expression between Bac-P and Bac-C but with the slope reversed, having higher expression in Bac-C than Bac-P (Fig. 4, F and G). This signifies that, while the overall analysis showed that Bac-C and Bac-P shared a set of common gene expression programs, they were differentiated in their predominant task. Their identities and activities seemed to be strongly correlated to their spatial distribution in the lobule, possibly establishing a gradient of cellular heterogeneity from the periphery to the center of the lobule.
We performed ISH analyses of the aquaporin AQPAe.a-like transcript that was identified as a marker of Bac-P (PE_Scaf12379_7.14) and found that, spatially, the transcripts were highly expressed by bacteriocytes in the periphery of the lobules (Fig. 4H). We further selected another Bac-P marker genes, cathepsins (PE_Scaf11249_3.4 and PE_Scaf11249_3.5), for ISH analysis to validate this. We showed that genes encoding both cathepsins were up-regulated in the lobule peripheral bacteriocytes (Fig. 4, I and J). Overall, the spatial distribution of these highly and uniquely expressed transcripts indicate that Bac-P cells were those bacteriocytes that were closer to the lobule periphery that may have better oxygen access from being closer to the peritoneum and efferent blood vessels; together with the function of the transcript-encoded protein products, Bac-P likely underwent aerobic respiration, nutrient transport, and processing of metabolic wastes. Another role of Bac-P may be to regulate symbiont abundance by actively degrading them intracellularly.
In contrast, Bac-C cells expressed a variety of genes encoding metabolic processing enzymes, including those involved in purine, carbohydrate, fatty acid, and amino acid metabolism (Fig. 4B and table S4), and notable among them are the SAM Mts. SAM is a universal biological cofactor found in all branches of life (48), and SAM Mts catalyze the transfer of methyl groups to various biomolecules, including DNA, proteins, and small-molecule secondary metabolites (49). FISH (Fig. 4K) and ISH (fig. S9, A and B) analyses of this essential marker gene further showed that genes encoding SAM Mts were primarily expressed in the cell nuclei of the bacteriocytes in the center of the lobules. These results suggest that Bac-C cells are those bacteriocytes located at the lobule center and thus may have less oxygen access. A similar expression pattern was also observed for key Bac-C markers, such as genes encoding phosphoenolpyruvate carboxykinase, glucose dehydrogenase, and l-sorbose 1-dehydrogenase (Fig. 4, L to N). While Bac-P also expressed these transcripts, they were much more prominently expressed in Bac-C and, based on the function of proteins encoded by these transcripts, we hypothesized that Bac-C is predominantly responsible for generating and processing symbiotic metabolic products (Fig. 4P).
Genes encoding hemerythrin and carbonic anhydrase, two crucial gas-transporting proteins, were also up-regulated in Bac-P in scRNA-seq data (Fig. 4B), which were the bacteriocytes located in the periphery of the lobules. Hemerythrin is an oxygen-binding protein found in various marine invertebrates (50), and carbonic anhydrase is an evolutionarily conserved carbon dioxide exchanger (51). Subsequent ISH analysis of these two transcripts differed from scRNA-seq results, showing both to be more highly expressed in the lobule center (Fig. 4O and fig. S7C). This inconsistency might be caused by the different environmental conditions between the two sampling sites of the two animals used for scRNA-seq and ISH; the tubeworm for scRNA-seq was collected from the F-site cold seep, while the tubeworms for FISH/ISH analysis were from the Haima seep. The Haima seep has a dissolved oxygen level of ~2 mg/liter (52), while the F-site has a higher level of around 3.2 mg/liter (53). The difference in oxygen levels may cause differences in the expression pattern of the oxygen-binding protein hemerythrin.
Summarily, our analysis of single-cell and spatial gene expression patterns suggest that Bac-P and Bac-C (Fig. 4P), while having common core bacteriocyte programming, may represent distinct subpopulations occupying different regions of the trophosome lobules, each supplying their respective symbionts with different gases and metabolites.
Symbionts’ morphologies and metabolic states are associated with microniches in trophosome
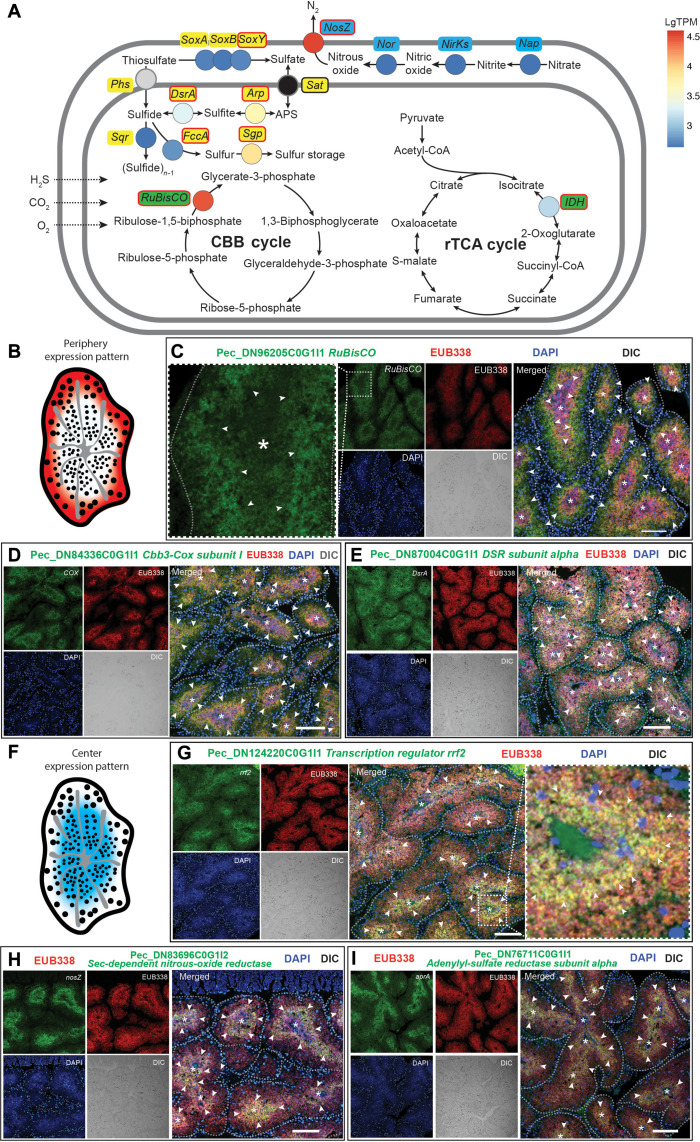
The trophosome microniches were associated with the symbionts’ morphological variations, with large symbionts residing in the lobule periphery and small symbionts in the lobule center (Fig. 1, C and D). It has been reported that the endosymbionts of P. echinospica and other siboglinid tubeworms have high metabolic versatility (26, 27). For example, both the gene encoding ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO), which controls the aerobic carbon fixation Calvin-Benson-Bassham (CBB) cycle (6, 41), and the gene encoding nitrous-oxide reductase (NosZ), which governs denitrification under anaerobic conditions, are known to be abundantly expressed in the symbionts of P. echinospica (27). The seemingly opposing metabolic states of the symbionts align well with the two bacteriocyte subpopulations described above as the Bac-P and Bac-C can respectively provide aerobic and anaerobic microinches for the symbionts. Therefore, we hypothesized that the symbionts’ metabolic states could be associated with their morphology (18) and corresponding bacteriocyte microniches. We analyzed the expression pattern of essential genes for the symbiont’s major metabolic pathways (sulfur, carbon, and nitrogen) to examine if their expression correlates with the symbionts’ morphology and microinches. These included essential genes of two sulfur metabolic pathways: the dissimilatory sulfite reductase (Dsr) pathway and the sulfite oxidation (Sox) pathway; the NosZ from the denitrification pathway; the aerobic carbon fixation CBB cycle; and the anaerobic carbon fixation rTCA cycle (Fig. 5A). The symbionts in the lobule periphery (Fig. 5B) strongly expressed genes that encode key enzymes in aerobic metabolic pathways (Fig. 5, C to E), including RuBisCO, DsrAB, and Cbb3-type cytochrome c oxidase subunit I (Cbb3-Cox I). In contrast, the symbionts in the lobule center (Fig. 5F) expressed genes that encode transcription regulator rrf2, NosZ, and adenylyl-sulfate reductase subunit alpha (AprA) (Fig. 5, G to I). We did not observe variation between the center and peripheral bacteriocytes in the expression of genes encoding sulfur storage protein, thiosulfate oxidation carrier protein (SoxY), and nicotinamide adenine dinucleotide phosphate–dependent isocitrate dehydrogenase (fig. S10).
Fig. 5. FISH analysis of metabolic states of the bacteriocytes in P. echinospica.
(A) Diagram showing the major metabolic pathways in the bacteriocytes. The color of each gene represents the expression level [lgTPM expression level based on (27)]. The genes labeled in red circles have been analyzed by FISH. (B) Diagram demonstrating the genes expressed in the periphery of a trophosome lobule. (C to E) Double FISH analyses of the key metabolic genes expressed in the symbionts located in the periphery of a trophosome lobule, including the genes encoding RuBisCO (Pec_DN96205C0G1I1), Cbb3-type cytochrome c oxidase subunit (Cbb3-Cox; Pec_DN84336C0G1I1), and Dsr alpha subunit (dsrA; Pec_DN96205C0G1I1). (F) Diagram demonstrating the genes expressed in the bacteriocytes in the center of a trophosome lobule. (G to I) Double FISH analyses of the key metabolic genes expressed in the symbionts in the center of a lobule, including the genes encoding transcription regulator rrf2 (rrf2; Pec_DN124220C0G1I1), sec-dependent NosZ (Pec_DN83696C0G1I2), and AprA (Pec_DN76711C0G1I1). The semitransparent dashed line marks the outline of each trophosome lobule, and the asterisk indicates the center of the lobules. The white arrowheads highlight regions exhibiting elevated expression of symbiont genes. The scale bars represent 100 μm.
Host-symbiont interaction in the periphery of lobules
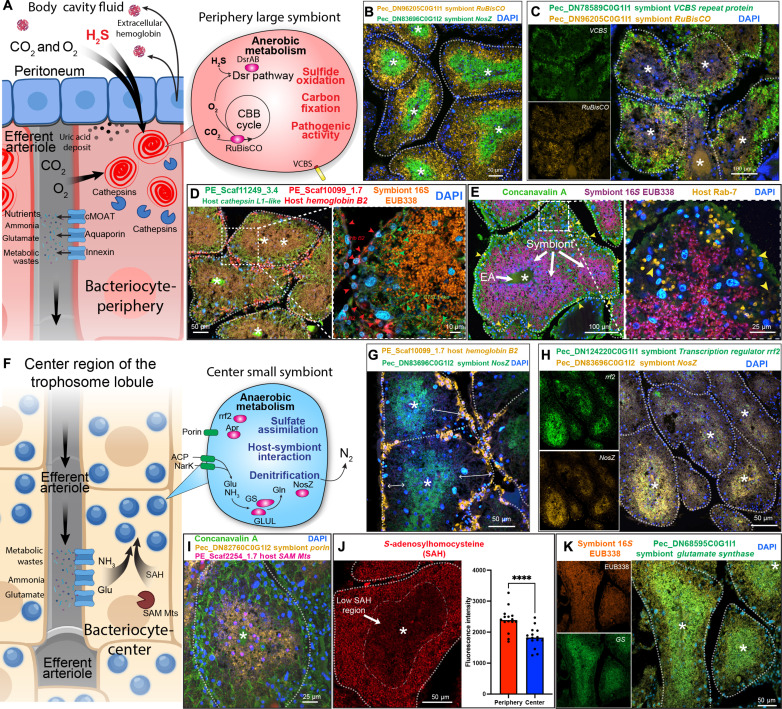
The joint interpretation of scRNA-seq data and image-based FISH/ISH analyses from both the host and the symbiont enabled further understanding of the host-symbiont interactions in P. echinospica symbiosis. Recent transcriptomic (19, 27, 38) and proteomic (18, 39) studies have revealed abundant expression of genes encoding gas transporters within the tubeworm trophosome. It has been proposed that a biochemical gradient is established by blood flow in the trophosome, leading to a differential concentration of metabolic substrates in different lobule regions (18). Our scRNA-seq data further suggest the presence of a biochemical gradient, transitioning from high-to-low concentrations from the periphery to the center of lobules within the P. echinospica trophosome. This gradient appears pivotal for facilitating gas exchange crucial for chemosynthesis (Fig. 6A). The peritoneum tissue of the trophosome synthesizes extracellular hemoglobin necessary for transporting oxygen and sulfide from the tubeworm’s plume and tail to the trophosome (Fig. 3B). These peritoneal cells synthesized the required protein subunits that are then presumably secreted into the coelomic cavity and assembled into complete extracellular hemoglobin (Fig. 6A). Physiologically, the peritoneum surrounds the lobule at the periphery (Fig. 3C); together, this evidence suggests that bacteriocytes residing in the lobule periphery bacteriocytes are in proximity to the site of extracellular hemoglobin production and circulation. Therefore, the periphery bacteriocytes may have access to a higher concentration of oxygen, carbon dioxide, and sulfide. This creates a microniche rich in chemosynthetic gases at the periphery. These peripheral symbionts can maintain aerobic respiration and active chemosynthesis, as marked by higher expression of gene encoding RuBisCO in a double FISH assay (Fig. 6B and fig. S11). In contrast, closer to the center of the lobule, oxygen and sulfide have been presumably consumed and were at lower concentrations. Therefore, symbionts within the center bacteriocytes likely conduct anaerobic respiration, marked by gene encoding NosZ in the same double FISH assay (Fig. 6B and fig. S10). In the periphery microniche, the symbiont can use the aerobic sulfite dissimilatory pathway to facilitate sulfide oxidation (Fig. 5E), which is further oxidized to sulfate, producing adenosine triphosphate (54, 55). Simultaneously, the elevated expression of RuBisCO in this microniche, which catalyzes the rate-limiting step in the CBB cycle, suggests that these periphery symbionts also fix atmospheric carbon dioxide into a biologically useful carbon source (17, 56).
Fig. 6. Host-symbiont interactions in P. echinospica symbiosis.
(A) Proposed model of host-symbiont interaction in the periphery of trophosome lobule. (B) Double FISH analysis of the genes encoding symbiont RuBisCO (Pezc_DN96205C0G1I1) and NosZ (Pec_DN83696C0G1I2), showing that RuBisCO and NosZ are expressed in the periphery and center symbionts, respectively. (C) Double FISH analysis of the genes encoding symbiont VCBS repeat protein (Pec_DN78589C0G1I1) and RuBisCO (Pec_DN96205C0G1I1), showing that both genes are expressed in periphery symbionts. (D) HCR analysis of the tubeworm host gene encoding cathepsin L1–like (PE_Scaf11249_3.4), host gene encoding hemoglobin B2 (PE_Scaf10099_1.7), and symbiont 16S rRNA. (E) FISH/IHC analysis of the symbiont 16S rRNA and late endosome marker Rab-7. The arrowheads indicate endosomes at the late stage of symbiont digestion. (F) Proposed mechanism of host-symbiont interaction in the center of a trophosome lobule. (G) Double FISH analysis of the tubeworm host gene encoding hemoglobin B2 (PE_Scaf10099_1.7) and symbiont gene encoding NosZ (Pec_DN83696C0G1I2); the double-sided arrows indicated the gaps between the NosZ-expressing anaerobic symbionts and the trophosome lobule’s peritoneum. (H) Double FISH analysis of the symbiont genes encoding transcription regulator rrf2 (Pec_DN124220C0G1I1) and NosZ (Pec_DN83696C0G1I2), showing that both genes are expressed in the lobule center symbionts. (I) Double FISH analysis of the tubeworm host gene encoding SAM-dependent methyl transferase (PE_Scaf2254_1.7) and the symbiont gene encoding porin (Pec_DN82760C0G1I2). (J) Fluorescence IHC analysis of SAH distribution in a trophosome lobule. The dashed-dotted line circled the low SAH region in the center of the lobule (P < 0.01, t test, n = 15). (K) FISH analysis of the symbiont 16S rRNA and the symbiont gene encoding glutamate synthase (Pec_DN68595C0G1I1), showing that the glutamate synthase is expressed in trophosome center symbionts.
Furthermore, the availability of gases can also regulate the pathogenicity of the symbiont. The RuBisCO-positive symbionts also expressed the gene encoding the VCBS repeat protein (fig. S12 and Fig. 6C). VCBS is one of the most abundantly expressed symbiont genes in the P. echinospica transcriptome (27). The VCBS domain-containing proteins are commonly found in pathogenic species of Vibrio, Colwellia, Bradyrhizobium, and Shewanella, hence the name VCBS (57). They have been proposed to be involved in bacterial adhesion and to play a role in virulence determination in pathogenic bacteria (58). The coexpression of RuBisCO and VCBS indicates that aerobic respiration could trigger the pathogenicity of the symbionts in the periphery microniche; oxygen-associated pathogenicity is common among various human gut pathogens (59).
Notably, symbiont bacteriocyte digestion is also associated with the periphery niche, which aligned well with previous ultrastructural and proteomics studies (9, 18). Figure 6D shows that the gene responsible for encoding cathepsin was mainly expressed in the bacteriocytes adjacent to the trophosome peritoneum, as identified by the expression of gene encoding B2 unit giant hemoglobin (fig. S13). Previous studies suggest that the host tubeworm uses endoplasmic reticulum–derived membrane/vacuoles and lysosomal activities as the primary mechanisms to digest symbionts (9, 18). To confirm this, we conducted a FISH/immunohistochemistry (IHC) analysis to examine if the endosomal and lysosomal marker RAB7 (60, 61) is associated with the periphery of the trophosome. Our findings indicate that the RAB7 signal was weak in the center bacteriocytes where the small symbionts reside; however, in the peripheral bacteriocytes, dual labeling clearly showed RAB7-positive membranes surrounding the large symbionts (Fig. 6E and fig. S14), indicating that the host could be using lysosomal vesicles to digest and extract nutrients from the bacteriocytes. In the outermost large RAB7-positive vesicles, the 16S FISH signal from the bacteriocytes was not detectable, suggesting that they were in the late stage of symbiont digestion approaching complete degradation.
Host-symbiont interactions in the center of the lobules
In the center of the trophosome lobules (Fig. 6F), the NosZ up-regulated symbionts are separated from the periphery niche (Fig. 6G), suggesting that symbionts in the center may mainly use nitrogen species but not oxygen as an electron acceptor. The gene encoding rrf2 transcription regulator, which is a widespread bacterial redox sensor (62), had been previously found abundantly expressed in the symbiont P. echinospica metatranscriptome (27). In the present study, the subsequent double FISH analysis of genes encoding rrf2 and NosZ showed that both genes were expressed in the center of the trophosome lobules with a strong positive correlation (r = 0.795). This suggests a regulatory relationship between the two and indicates that the lobule center is a hypoxic niche. In addition, the gene encoding AprA is highly expressed in the center symbionts (Fig. 5I). The adenylyl-sulfate reductase enzyme plays a crucial role in the evolutionarily conserved adenosine 5′-phosphosulfate pathway that regulates the rate-limiting step of sulfur assimilation and dissimilation by reversible converting inorganic sulfate to cysteine (63, 64). Thus, the elevated expression of AprA by center symbionts could be related to a lower sulfide concentration in this microniche, possibly because most of the sulfide has already been consumed by periphery symbionts.
On the basis of the results above, the center symbionts may have to obtain sulfur species from the host through host-symbiont metabolite interactions. In R. pachyptila, proteins involved in host-symbiont interactions were mainly up-regulated in the small symbionts in the center of the lobules (18). The porin protein, which is one of the most abundantly expressed small symbiont proteins in R. pachyptila (18), is particularly intriguing because it has a crucial role in shuffling metabolites between the host animal and microbe (65, 66). Previous bulk metatranscriptome analysis also reported that the gene encoding porin was abundantly expressed in the symbiont of P. echinospica (27). Using FISH analysis, the P. echinospica symbiont porin was found to be expressed highly in the center symbionts (fig. S15), suggesting an exchange of metabolites between the host and symbionts in the center of the lobules.
The scRNA-seq results demonstrated that the bacteriocytes in the center of the lobules produced symbiont-required metabolites and niche-specific enzymes that may be involved in host-symbiont interaction (Fig. 4B). Most noticeably, Bac-C expressed several SAM Mts (Fig. 4K and fig. S8, A and B). A double FISH analysis shows that the SAM Mts-expressing bacteriocytes overlap with the porin-positive symbionts (Fig. 6I and fig. S16), further suggesting the host SAM Mts are associated with host-symbiont interaction. The substrate (SAM) and metabolic product [S-adenosylhomocysteine (SAH)] of the SAM Mts may be crucial for such interactions. One possibility is that SAM and SAH can act as autoinducers in bacterial quorum sensing, which might be involved in regulating the cell density of bacterial biofilms (67). Furthermore, SAH can be broken down by SAH hydrolase into l-homocysteine, for which the symbiont can either convert to methionine through remethylation or to cystathionine through the transsulfuration pathway (68). The SAH distribution in the trophosome lobules, as revealed by in situ fluorescence IHC analysis, shows that the presence of SAH was higher in the periphery of the lobules than in the center (Fig. 6J and fig. S17). This finding contradicts the elevated expression of host genes encoding SAM Mts in the center of the lobules. We suspect that the symbionts had already consumed the SAH produced by the bacteriocytes in the center of the lobules.
The center of trophosome lobules harness an ammonia detoxification mechanism
Our results suggest that the tubeworm host actively produces metabolites to support the sulfur-driven denitrification of symbionts in the center of trophosome lobules but does not consume these symbionts as a nutrient source. We then ask how the host tubeworm benefits from this unique arrangement. The trophosome generates nitrogenous metabolic waste, like urea and ammonia, through the digestion of symbionts and nutrient metabolism (69). To address the toxicity of these nitrogenous metabolites, the tubeworm must have developed mechanisms to mitigate their effects. In the present study, we found an up-regulation of the gene encoding xanthine dehydrogenase/oxidase in bacteriocytes, which is an enzyme that converts hypoxanthine to xanthine and xanthine to urate, the final step in purine catabolism (70). We also observed high amounts of urate granules in the trophosome lobules’ periphery using Gomori methenamine silver staining (fig. S18). Our findings are consistent with previous studies on R. pachyptila, indicating that the periphery of the tubeworm trophosome lobules can convert nitrogenous wastes into urate or uric acid deposits (13), a less toxic form that symbionts might use as a bioavailable source of nitrogen (19).
The host tubeworm must also process ammonia derived from amino acid metabolism (71). The genes encoding ammonia transporters, such as AQPs (72, 73), were up-regulated in the Bac-P and the extracellular vesicles (EVs), indicating that the ammonia produced by the Bac-P is excreted into the blood and further transported inward to the lobule center. We posit that the lobule center has an ammonia detoxification mechanism maintained by both the host and symbionts as symbionts in the lobule center are actively conducting denitrification. To determine if these center symbionts are capable of transporting and detoxifying ammonia by incorporating ammonia into its nitrogen metabolism, we examined the expression of the gene encoding symbionts’ ammonia transporter (ammonia channel protein, Pec_DN101659C0G1I1) and genes involved in ammonia assimilation: glutamine synthase (Pec_DN68595C0G111) and glutamate-ammonia ligase (Pec_DN115499C0G1I1). Our results show that these genes were expressed in the center symbionts (Fig. 6K and fig. S19), which suggests that the center symbionts can absorb and detoxify ammonia.
Perspectives and limitations
The gutless vestimentiferan tubeworm is a well-known species from the deep-sea chemosynthetic ecosystem. However, we still need to better understand the mechanisms used by the tubeworm to interact with and regulate its endosymbionts. In the present study, we investigated the specific functions of different trophosome cell types in the deep-sea tubeworm P. echinospica, using a combination of deep-sea in situ single-cell sample fixation, cell FACS sorting, and scRNA-seq to profile the transcriptomes of individual trophosome cells. Our results show that the trophosome has specific cell types expressing genes encoding for proteins that can transport gases and shuttle metabolites. These cells appear to create a high-to-low biochemical gradient for fueling substrates of chemosynthesis from the periphery to the center of the trophosome lobule. We identified two bacteriocyte populations representing two distinct micro-niches within each trophosome lobule. Integrated analysis of the symbiont’s key metabolic pathways indicates an association between the metabolic status of the bacteriocytes and their locations in the lobules. The bacteriocytes in the oxygen-rich peripheral niche of the lobules actively conduct autotrophic carbon fixation. In contrast, the symbionts in the hypoxic center niche of the lobules conduct anaerobic denitrification, helping the host to remove ammonia wastes. By coordinating endosymbiotic activity, the trophosome can produce and process chemosynthetically fixed nutrients while storing and detoxifying metabolic wastes. Ultimately, it will be important to identify the nutrients and metabolites that control the biological and morphological guardians of the endosymbionts. However, neither the P. echinospica nor its endosymbiont can be cultured under laboratory conditions, posing a challenge to directly assess the intratissue concentration of chemosynthetic gases like hydrogen sulfide and oxygen. Furthermore, characterizing the function of key genes directly through loss-of-function techniques like RNA interference, morpholino, or CRISPR-Cas editing remains extremely difficult and impractical for these deep-sea animals living in the cold seep. Despite these limitations, the single-cell atlas developed in the present study begins to shed light on the common principles of symbiosis and environmental adaption of animals in the deep sea at a molecular level. The workflow developed in the present study also represents a new experimental paradigm to generate such molecular-level characterizations for investigating biological adaptation in important nonmodel animals.
MATERIALS AND METHODS
In situ fixation for scRNA-seq
HD-ACME [modified from ACME solution reported by García-Castro et al. (40)] solution was freshly prepared using a 3:3:2:12 ratio of deoxyribonuclease (DNase)/ribonuclease (RNase)-free milliQ water, methanol, glacial acetic acid, and glycerol. We adjusted the portions of glycerol to increase the density of the original ACME solution. The HD-ACME solution has a higher density than the deep-sea water and, therefore, would stratify with deep-sea water during sampling. Five liters of HD-ACME solution was filled into the deep-sea animal fixation apparatus (fig. S2). In addition, 50 ml of 7.5% N-acetyl-l-cysteine (NAC) solution was added to each fixation apparatus before each ROV dive.
At the F-site cold seep, one P. echinospica tubeworm was collected (GPS location 22°06′5824″N, 119°17′086″E; depth of ~1187.3 m). The tubeworm was picked up, slightly twisted, and cracked open using the mechanical arm of the ROV Faxian and transferred into the fixation apparatus filled with HD-ACME solution. The fixation apparatus was then sealed and retrieved with the ROV.
Once onboard R/V Kexue, the tubeworm was quickly put on ice. The trophosome of the tubeworm was carefully dissected and transferred to 15-ml Falcon tubes containing 10 ml of ice-cold regular ACME solution (13:3:2:2 ratio of DNase/RNase-free milliQ water, methanol, and glacial acetic acid, supplemented with 100 μl of 7.5% NAC). The tube was kept on ice and gently shaken on a seesaw shaker at 30 rpm for 45 min. The samples were gently pipetted up and down several times to complete dissociation using 1-ml pipette tips. We then centrifuged the samples at 1000g for 10 min (4°C) and discarded the supernatant ACME solution. Each pellet was resuspended in 900 μl of resuspension buffer [1× Dulbecco’s phosphate-buffered saline (PBS) and 1% bovine serum albumin (BSA)] and transferred to RNase-free cryogenic vial (Thermo Fisher Scientific). Then, 100 μl of dimethyl sulfoxide was added to each vial and mixed well. The vials were stored at −80°C.
Tubeworm sample collection
The P. echinospica tubeworms for experiments other than scRNA-seq were collected from Haimai seep during the R/V Haiyangdizhi 6 2021 and 2022 cruise (cruise numbers: HYDZ6-202102 and HYDZ6-202203) by ROV Haima. Once onboard R/V Haiyangdizhi 6, the tubeworms were quickly dissected on ice. The worms were then fixed in prechilled 4% paraformaldehyde (PFA) prepared in filtered and autoclaved in situ seawater collected from previous ROV dives. After fixing overnight (about 14 to 16 hours), the tubeworms were washed three times in ice-cold RNase-free PBS. The specimens were then dehydrated and stored in 100% methanol at −20°C.
FACS and library construction
To prepare the cells for scRNA-seq, we used a combination of FACS and library construction using the 10X protocol (Chromium Next GEM Single Cell 3′ Reagent Kits v3.1). ACME-dissociated and fixed cells were first filtered through 40-μm pluriStrainer strainers and then stained with DRAQ5 nuclear dye (eBioscience) and concanavalin A conjugated with Alexa Fluor 488 (Invitrogen) to label the cytoplasm. The stained cells were visualized using a CytoFlex S Flow Cytometer (Beckman Coulter) or sorted using a BD Influx cell sorter. To avoid RNAse contamination during cell sorting, the FACS instrument was thoroughly decontaminated with bleach, and sorting was performed using the BD FACSDiva Software in 4-Way Purity mode with a 70-μm nozzle and moderate-pressure separation (65 psi). We sorted DRAQ5-positive, concanavalin A–positive single cells and collected them in 300-μl tubes containing 5 μl of collection buffer (1× PBS and 1% BSA) and 1 μl of RNAse inhibitor (1× PBS and 1% BSA), obtaining up to 50,000 cells per tube. Three libraries were constructed according to the Chromium Next GEM Single Cell 3′ Reagent Kits v3.1 protocol. Nine microliters of trypan blue was added into 1 μl of cell suspension and loaded into Countess II FL Automated Cell Counter. The volume of the cell suspension added into the master mix of 10X depending on the cell concentration and the number of targeted cell recovery. In our experiment, 16.5 μl of cell suspension was added into the master mix and loaded with beads and partitioning oil into the microfluidics machine, then after the formation of GEM generation and barcoding, followed by GEM-RT, cDNA amplification, and cDNA quality control and quantification. Last, according to the concentration of cDNA, the library was constructed according to the fragmentation, end repair, A-tailing, size selection, and sample index polymerase chain reaction (PCR) process in the original protocol. The libraries were then sequenced with the Illumina HiSeq 4000 platform.
scRNA-seq data analysis
Three sets of scRNA-seq libraries were generated to analyze the trophosome in P. echinospica. To map the single-cell transcriptome data, gtf annotation files for both the host and symbiont are needed. To generate the required gtf files, we used the TAR-seq pipeline developed by Wang et al. (74), which generates a gft of the coordinates of transcriptionally active regions (TARs), generated using RNA-seq data. Briefly, in well-annotated genomes, these TARs correspond to annotated gene regions (aTARs), but for less well-annotated genomes, these TARs also include “transcriptional dark matter,” which is observed to be transcribed but may not be annotated (uTARs). The generation of TARs enables annotation-free analysis of scRNA-seq data, which we apply here due to the lack of symbiont reference annotation gtf. The TAR algorithm took the symbiont reference genome (27) and scRNA-seq data as input and generated a gtf file of TARs in the symbiont. The genome fasta for the host, symbiont, and host mitochondria were respectively obtained from previously published studies (27, 38, 75) and were merged to create a combined genome fasta file. Along with both the host gtf of gene annotations and symbiont gtf of TARs, we created the combined host/symbiont reference using the CellRanger MakeRef tool (v6.1.2 with default settings). The host fasta file contains 14 super scaffolds, and gtf file contains 22,642 annotated genes (38). The symbiont genome fasta file comprised 15 long scaffolds (27), and 2306 TARs were identified by the TAR-seq pipeline.
After creating this custom host/symbiont reference, we mapped the scRNA-seq data simultaneously to the host and symbiont, generating a matrix of host gene/symbiont TAR expression (fig. S20). Three scRNA-seq libraries from three technical replicates were mapped against our custom reference genome using CellRanger Count (v6.1.2; parameter: --expect-cells=4000). Genome mapping rates for these libraries are 86.3, 88.4, and 89% for three technical replicates; in addition, 57.8, 56.3, and 56.6% reads are confidently mapped to the genome, and 33.5, 36.5, and 36.9% are confidently mapped to the transcriptome. After removing symbiont reads, each cell has a median unique molecular identifiers (UMI) count of 411 and a median gene count of 227; before filtering symbiont reads, the mean UMI count per cell was 666, and the mean gene count per cell was 239. Across all 5385 pass-QC cells, we detected 10,745 P. echinospica genes of the 20,733 protein-coding genes in the genome that have matched functional annotations (38). While the number of genes and UMIs detected in scRNA-seq on model organisms is typically higher, these scRNA-seq metrics are comparable to that of other published reports on marine invertebrates such as Xenia sp. (freshly dissociated; median UMI per cell, 801; median genes per cell, 467) (76) and Nematostella (García-Castro et al.: ACME fixed; median UMI per cell, 671; median genes per cell, 418; Sebé-Pedrós et al.: freshly dissociated; median UMI per cell, 541; median genes per cell, 278) (40, 77). Freshly dissociated samples are generally of higher quality and produce more UMIs and genes detected than fixed samples. The P. echinospica samples used in our study are obtained from deep-sea cold seeps and fixed in situ using a modified HD-ACME fixative (40), and the necessary use of this fixative may also contribute to lower UMI and gene detection numbers. As it is not possible to obtain fresh samples for this deep-sea animal, we anticipate that future advances and optimizations using our method as a starting point will further improve the quality of scRNA-seq data of deep-sea animals.
The resulting gene expression matrices, representing cell count data, were analyzed using R v4.1.2 with the Seurat v4.3.0.1 package (78). The three datasets were integrated using Seurat, and cells were considered for further analysis based on specific filtering criteria. We require the RNA counts (nCount_RNA) to be greater than 600 but less than 5000, and the percentage of mitochondrial gene expression (percent.mt) to be between 30 and 80. These two criteria together can ensure removal of empty droplets that only contain contaminant mitochondrial reads from lysed cells while keeping viable cells that may have high mitochondrial expression.
The count matrix underwent standard preprocessing steps in Seurat. Upon examination of the datasets, it became evident that the expression of symbiont genes permeates a diverse array of cell types. Thus, in our downstream analysis, only the host gene matrices are filtered for normalization and downstream clustering. Counts were normalized and log-scaled using the NormalizeData and ScaleData functions, respectively. Principal components analysis (PCA) was performed (RunPCA) using only highly variable features (FindVariableFeatures). The top 20 principal components were selected based on an elbow plot of standard deviation. Clustering was performed with a resolution of 0.15 using the Seurat FindClusters function. Marker genes were identified using the FindAllMarkers function, considering only genes enriched and expressed in at least 25% of cells in each population (min.pct = 0.25) with a log(fold difference) larger than 0.25 (logfc.threshold = 0.25).
We used cluster markers for GO enrichment analysis. The ClusterProfiler (79) R package was used to determine statistically significant differential gene expression in GO terms of molecular function, cellular component, and biological process.
Trajectory analysis of bacteriocytes (Bac-C and Bac-P) was performed using monocle 3’s (80, 81) learn_graph function to estimate cellular ordering and connectivity. On the basis of the previous literature and the assessment of gene expression markers such as SAM, aquaporin, and hemoglobin through FISH, we hypothesized that Bac-C cells represent an earlier stage, while the peripheral Bac-P cells correspond to a later stage in the developmental process. Consequently, we designated the Bac-C cells as the origin of the pseudotime trajectory expression. Because there are only two cell types present, the direction of the trajectory, regardless of whether it is toward Bac-C or Bac-P cells, will provide valuable insights into the observed gradient of gene.
H&E, FISH, and IHC analysis of the trophosome
The P. echinospica truck segments, which contain the trophosome, were embedded in Epredia histoplast paraffin (Thermo Fisher Scientific, MA, United States) using a Revos tissue processor (Thermo Fisher Scientific) with the 13-hour general program [70, 75, 90, 95, and 3× 100% ethanol washes, 1 hour each at room temperature (RT); 3× xylene washes, 1 hour each at RT; and 3× paraffin washes, 1 hour each at 62°C]. The specimens were then mounted with a HistoStar embedding workstation (Thermo Fisher Scientific). Tissue sections (5 μm in thickness) were cut using HM325 microtome (Thermo Fisher Scientific). For H&E stains, tissue sections were dewaxed with a standard protocol (3× xylene washes; xylene:ethanol 1:1 wash; 3× 100% ethanol washes; 95, 85, 75% ethanol washes; all 5 min each at RT). The H&E staining and Gomori methenamine silver staining were conducted with a standard H&E histology staining kit and urate stain kit, respectively (SolarBio, Beijing, China).
For FISH/IHC analysis, the tissue sections were dewaxed and then washed three times with PBST (1× PBS and 0.1% Tween 20) for 5 min at RT. Then, the sections were hybridized with the digoxigenin-labeled EUB338 (82) probe according to the FISH method described above. After washing, the slides were blocked with blocking buffer (2% BSA and 2% sheep serum) for 1 hour at RT. Then, the slides were incubated with 1:2500 diluted anti–Dig-peroxidase (POD) antibody (Roche) and 1:200 diluted late endosome marker anti–Rab-7 rabbit antibody (Abcam) or 1:200 diluted anti-SAH (S-(5′-adenosyl)-l-homocysteine) rabbit antibody (Sangon, Shanghai) overnight at 4°C. The slides were washed five times with PBST for 15 min each at RT. The 16S FISH signal was first developed using the Cy5 TSA fluorescein kit (Akoya Biosciences) according to the manufacturer’s protocol. After three 5-min PBST washes, the slides were incubated with 1:1000 Alexa Fluor 555–labeled donkey anti-rabbit secondary antibody (Thermo Fisher Scientific) for 1 hour at RT. After another three 5-min PBST washes, the slides were stained with 4′,6-diamidino-2-phenylindole (DAPI) and Alexa Fluor 488 conjugate concanavalin A and lastly mounted and imaged with the abovementioned method.
Colorimetric ISH
The DNA fragments (~1000 base pairs) of the targeted genes were first PCR amplified using P. echinospica trophosome cDNA (for host genes) or trophosome genomic DNA (for symbiont genes). The amplified fragments were ligated into the pMD19-T vector (Takara) and transformed into Escherichia coli. Individual colonies were picked up, and their plasmids were sequenced to confirm the inserts. The in vitro mRNA transcription templates were amplified using T7 forward GSP (sense probe control) or Sp6 reverse GSP (antisense probe) primers (table S5). The digoxigenin- or fluorescein-labeled antisense probes and sense control probes were generated according to the method described by Thisse and Thisse (83).
For colorimetric ISH, the tissue sections were dewaxed according to the abovementioned method. The tissue sections were washed three times with PBST for 5 min each at RT and then permeabilized with proteinase K (2 μg/ml) in PBST for 30 min at 37°C. The postdigestion fixation was conducted by fixing the tissue sections with 4% PFA in PBST for 30 min at RT. After 3× 5-min PBST washes to remove the residual fixative, the tissue sections were prehybridized with hybridization mix (HM) [containing 50% formamide, 5× saline-sodium citrate (SSC), 0.1% Tween 20, heparin (10 μg/ml), and yeast tRNA (500 μg/ml)] for 1 hour at 65°C. Then, the prehybridization HM was removed, and the tissue section was incubated in fresh HM containing Dig-labeled probe (~0.5 ng/μl). The hybridization was conducted in a moisture chamber in a 55°C incubator overnight. Posthybridization washes were performed according to the following steps: The tissue sections were first washed 3× for 15 min with 2× SSC with 0.1% Tween 20 and then 3× for 15 min with 0.2× SSC with 0.1% Tween 20. After cooling down to RT, the tissue sections were washed 3× for 5 min with PBST and then incubated in blocking buffer for 1 hour at RT. The tissue sections were incubated with 1:2500 diluted anti–Dig-AP antibody (Roche) at 4°C overnight. The tissue sections were washed 6× for 15 min with PBST, followed by washing 3× for 15 min with alkaline Tris buffer [100 mM tris-HCl (pH 9.5), 100 mM NaCl, and 50 mM MgCl2]. The samples were incubated in nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate staining solution (Abcam). After the desired expression pattern was revealed, the staining reaction was stopped by 3× 15-min PBST-EDTA wash (PBST and 1 mM ETDA). The tissue sections were then mounted with 80% glycerol. For each gene, we also conducted control hybridization (with the sense probe) and found no obvious signal.
Multicolor fluorescent ISHs
For double ISH analysis, the prehybridized tissue sections were hybridized with HM containing Dig- and fluorescein-labeled probes. The hybridization, posthybridization washes, and blocking were conducted using the methods described above. Then, the sections were first incubated with 1:1000 diluted anti–fluorescein-POD (Roche) overnight at 4°C and then washed six times with PBST for 15 min each and three times with TNT buffer [100 mM tris-HCl (pH 7.5), 100 mM NaCl, and 0.1% Tween 20] for 15 min each. Afterward, the fluorescent signal of fluorine-labeled probes was developed by TSA fluorescein kit (Akoya Biosciences) according to the manufacturer’s protocol. The slices were then washed three times, and the remaining POD activity was quenched by incubation in 1% hydrogen peroxide solution for 1 hour at RT. Then, the slices were washed three times with PBS, blocked with blocking buffer for 30 min at RT, incubated with 1:2500 diluted anti–Dig-POD (Roche) overnight at 4°C, and washed six times with PBST and three times with TNT. The signal was developed using the TSA Cy3 kit (Akoya Biosciences). Last, the slices were washed with PBST, stained with DAPI, and mounted with ProLong Glass.
For hybridization chain reaction (HCR)/ISH analysis, the probes and amplifier were ordered from Sangon Biotech (Sangon, Shanghai), and their sequences were provided in table S6. The HCR/ISH analysis was conducted according to the methods described by Choi et al. (84). The section slices were first dewaxed and rehydrated according to the abovementioned methods. After washing with PBST for 15 min at RT, the slices were prehybridized in 50 μl of HCR hybridization buffer for 30 min at 37°C, which was then replaced with 50 μl of HCR hybridization buffer with 8 nM each of the probes. The slices were incubated at 37°C overnight. The following day, the sections were washed five times with HCR probe wash solution (preheated to 37°C, 300 μl each time) at 37°C and then three times in 1 ml of 5× SSCT for 5 min at RT. The sections were preamplified in 100 μl of amplification buffer for 30 min at RT. At the same time, 2 μl of each amplifier hairpin (for three probe B1–H1–Alexa Flour 488 labeled, B1–H2–Alexa Fluor 488 labeled, B2-H1-Atto550 labeled, B2-H2-Atto550 labeled, B3–H1–Alexa Fluor 647 labeled, and B3–H2–Alexa Fluor 647 labeled) per section slice was placed in separate thin PCR tubes. The amplifiers were denatured in a PCR thermocycler for 90 s at 95°C and cooled to RT in the dark for 30 min. Then, all hairpins were pooled in one tube with 50 μl per section of amplification buffer. Then, 50 μl of amplification buffer and the hairpin mix was added to each section and incubated in the dark overnight at RT. The next day, the sections were washed three times in 5× SSCT for 10 min at RT. Last, the slices were stained with DAPI and mounted with ProLong Glass.
Microscopy imaging and image processing
All the colorimetric ISH images were observed and imaged with a Nikon Eclipse Ni microscope. All the fluorescent FISH/ISH slides were imaged with a Nikon Ax or a Zeiss LSM900 confocal microscope. Quantification of mean fluorescence intensity was performed in ImageJ.
Reproducibility
We first conducted colorimetric ISH on at least three tubeworms to examine the expression pattern of markers of peritoneal cells, EA cells, bacteriocytes, and symbionts. The expression patterns were consistent across all the individuals. The double FISH, multicolor FISH/HCR, and FISH/IHC analyses were also repeated in three individuals to confirm the consistency of the expression pattern. We conducted colorimetric ISH on a single tubeworm specimen for the cluster markers of spermatozoa cells.
Acknowledgments
Funding: This work was supported by grants from the Science and Technology Innovation Project of Laoshan Laboratory (project no. LSKJ202203104), National Natural Science Foundation of China (grant no. 42030407), the Guangdong Government (2019B030302004), Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou) (2021HJ01 and SMSEGL20SC01), the HKSAR government (16101822 and C2013-22G), and a special research donation from the Chau Hoi Shuen Foundation.
Author contributions: H.W., A.R.W., C. Li, and P.-Y.Q. designed the study. C. Lian and H.W. designed the in situ animal fixation apparatus. H.W., G.Y., H.Z., C. Lian, Z.Z., and M.W. collected the tubeworm samples. X.X., B.F., Y.L., C.W.F., and A.R.W. performed the bioinformatics analysis. H.W., X.X., H.Z., Z.Z., and J.L. performed the experiments. H.W., X.X., A.R.W., C. Li, and P.-Y.Q. wrote this paper. All authors revised the manuscript. All authors read, approved, and contributed to the final manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The raw sequencing data of trophosome scRNA-seq have been deposited in the National Center for Biotechnology Information (NCBI) Short Read Archive under the accession number PRJNA1086801. The RDS object was uploaded to Figshare (doi.org/10.6084/m9.figshare.25332232). The genome, transcriptome, and annotation of P. echinospica are available at the NCBI under BioProject PRJNA625616 and Figshare (doi.org/10.6084/m9.figshare.15050478). The mitochondria genome of P. echinospica is available at GenBank under accession number MG462707. The P. echinospica symbiont genome and transcriptome are available at GenBank under accession numbers RZUD00000000 and GHDL00000000, respectively. All analysis codes for scRNA-seq are available at Figshare (doi.org/10.6084/m9.figshare.25332232).
Supplementary Materials
This PDF file includes:
Figs. S1 to S20
Legends for tables S1 to S6
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S6
REFERENCES AND NOTES
- 1.Jones M. L., Riftia pachyptila Jones: Observations on the vestimentiferan worm from the Galápagos Rift. Science 213, 333–336 (1981). [DOI] [PubMed] [Google Scholar]
- 2.Southward E. C., Schulze A., Tunnicliffe V., Vestimentiferans (Pogonophora) in the Pacific and Indian Oceans: A new genus from Lihir Island (Papua New Guinea) and the Java Trench, with the first report of Arcovestia ivanovi from the North Fiji Basin. J. Nat. Hist. 36, 1179–1197 (2002). [Google Scholar]
- 3.Boetius A., Microfauna-macrofauna interaction in the seafloor: Lessons from the tubeworm. PLOS Biol. 3, e102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rouse G. W., Goffredi S. K., Vrijenhoek R. C., Osedax: Bone-eating marine worms with dwarf males. Science 305, 668–671 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Cavanaugh C. M., Gardiner S. L., Jones M. L., Jannasch H. W., Waterbury J. B., Prokaryotic cells in the hydrothermal vent tube worm Riftia pachyptila Jones: Possible chemoautotrophic symbionts. Science 213, 340–342 (1981). [DOI] [PubMed] [Google Scholar]
- 6.Felbeck H., Jarchow J., Carbon release from purified chemoautotrophic bacterial symbionts of the hydrothermal vent tubeworm Riftia pachyptila. Physiol. Zool. 71, 294–302 (1998). [DOI] [PubMed] [Google Scholar]
- 7.Markert S., Arndt C., Felbeck H., Becher D., Sievert S. M., Hugler M., Albrecht D., Robidart J., Bench S., Feldman R. A., Hecker M., Schweder T., Physiological proteomics of the uncultured endosymbiont of Riftia pachyptila. Science 315, 247–250 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Hand S. C., Trophosome ultrastructure and the characterization of isolated bacteriocytes from invertebrate-sulfur bacteria symbioses. Biol. Bull. 173, 260–276 (1987). [DOI] [PubMed] [Google Scholar]
- 9.Monika B., Angelika S., Ultrastructural reinvestigation of the trophosome in adults of Riftia pachyptila (Annelida, Siboglinidae). Invertebr. Biol. 122, 347–368 (2003). [Google Scholar]
- 10.Arp A. J., Doyle M. L., Di Cera E., Gill S. J., Oxygenation properties of the two co-occurring hemoglobins of the tube worm Riftia pachyptila. Respir. Physiol. 80, 323–334 (1990). [DOI] [PubMed] [Google Scholar]
- 11.Zal F., Suzuki T., Kawasaki Y., Childress J. J., Lallier F. H., Toulmond A., Primary structure of the common polypeptide chain b from the multi-hemoglobin system of the hydrothermal vent tube worm Riftia pachyptila: An insight on the sulfide binding-site. Proteins 29, 562–574 (1997). [PubMed] [Google Scholar]
- 12.De Cian M.-C., Andersen A. C., Bailly X., Lallier F. H., Expression and localization of carbonic anhydrase and ATPases in the symbiotic tubeworm Riftia pachyptila. J. Exp. Biol. 206, 399–409 (2003). [DOI] [PubMed] [Google Scholar]
- 13.De Cian M., Regnault M., Lallier F. H., Nitrogen metabolites and related enzymatic activities in the body fluids and tissues of the hydrothermal vent tubeworm Riftia pachyptila. J. Exp. Biol. 203, 2907–2920 (2000). [DOI] [PubMed] [Google Scholar]
- 14.Minic Z., Herve G., Arginine metabolism in the deep sea tube worm Riftia pachyptila and its bacterial endosymbiont. J. Biol. Chem. 278, 40527–40533 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Cordes E. E., Arthur M. A., Shea K., Arvidson R. S., Fisher C. R., Modeling the mutualistic interactions between tubeworms and microbial consortia. PLOS Biol. 3, e77 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Powell M. A., Somero G. N., Blood components prevent sulfide poisoning of respiration of the hydrothermal vent tube worm Riftia pachyptila. Science 219, 297–299 (1983). [DOI] [PubMed] [Google Scholar]
- 17.Elsaied H., Kimura H., Naganuma T., Molecular characterization and endosymbiotic localization of the gene encoding D-ribulose 1,5-bisphosphate carboxylase-oxygenase (RuBisCO) form II in the deep-sea vestimentiferan trophosome. Microbiology (Reading) 148, 1947–1957 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Hinzke T., Kleiner M., Meister M., Schlüter R., Hentschker C., Pané-Farré J., Hildebrandt P., Felbeck H., Sievert S. M., Bonn F., Völker U., Becher D., Schweder T., Markert S., Bacterial symbiont subpopulations have different roles in a deep-sea symbiosis. eLife 10, e58371 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Oliveira A. L., Mitchell J., Girguis P., Bright M., Novel insights on obligate symbiont lifestyle and adaptation to chemosynthetic environment as revealed by the giant tubeworm genome. Mol. Biol. Evol. 39, msab347 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lutz R. A., Shank T. M., Fornari D. J., Haymon R. M., Lilley M. D., Von Damm K. L., Desbruyeres D., Rapid growth at deep-sea vents. Nature 371, 663–664 (1994). [Google Scholar]
- 21.McClain C. R., Balk M. A., Benfield M. C., Branch T. A., Chen C., Cosgrove J., Dove A. D., Gaskins L., Helm R. R., Hochberg F. G., Lee F. B., Marshall A., McMurray S. E., Schanche C., Stone S. N., Thaler A. D., Sizing ocean giants: Patterns of intraspecific size variation in marine megafauna. PeerJ 3, e715 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lelièvre Y., Sarrazin J., Marticorena J., Schaal G., Day T., Legendre P., Hourdez S., Matabos M., Biodiversity and trophic ecology of hydrothermal vent fauna associated with tubeworm assemblages on the Juan de Fuca Ridge. Biogeosciences 15, 2629–2647 (2018). [Google Scholar]
- 23.Wang X., Guan H., Qiu J.-W., Xu T., Peckmann J., Chen D., Feng D., Macro-ecology of cold seeps in the South China Sea. Geosyst. Geoenviron. 1, 100081 (2022). [Google Scholar]
- 24.Gardebrecht A., Markert S., Sievert S. M., Felbeck H., Thurmer A., Albrecht D., Wollherr A., Kabisch J., Le Bris N., Lehmann R., Daniel R., Liesegang H., Hecker M., Schweder T., Physiological homogeneity among the endosymbionts of Riftia pachyptila and Tevnia jerichonana revealed by proteogenomics. ISME J. 6, 766–776 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reveillaud J., Anderson R., Reves-Sohn S., Cavanaugh C., Huber J. A., Metagenomic investigation of vestimentiferan tubeworm endosymbionts from Mid-Cayman Rise reveals new insights into metabolism and diversity. Microbiome 6, 19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y., Liles M. R., Halanych K. M., Endosymbiont genomes yield clues of tubeworm success. ISME J. 12, 2785–2795 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Y., Sun J., Sun Y., Kwan Y. H., Wong W. C., Zhang Y., Xu T., Feng D., Zhang Y., Qiu J. W., Qian P. Y., Genomic, transcriptomic, and proteomic insights into the symbiosis of deep-sea tubeworm holobionts. ISME J. 14, 135–150 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Polzin J., Arevalo P., Nussbaumer T., Polz M. F., Bright M., Polyclonal symbiont populations in hydrothermal vent tubeworms and the environment. Proc. Biol. Sci. 286, 20181281 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Markert S., Gardebrecht A., Felbeck H., Sievert S. M., Klose J., Becher D., Albrecht D., Thurmer A., Daniel R., Kleiner M., Hecker M., Schweder T., Status quo in physiological proteomics of the uncultured Riftia pachyptila endosymbiont. Proteomics 11, 3106–3117 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Arp A. J., Childress J. J., Sulfide binding by the blood of the hydrothermal vent tube worm Riftia pachyptila. Science 219, 295–297 (1983). [DOI] [PubMed] [Google Scholar]
- 31.Childress J. J., Arp A. J., Fisher C. R., Metabolic and blood characteristics of the hydrothermal vent tube-worm Riftia pachyptila. Mar. Biol. 83, 109–124 (1984). [Google Scholar]
- 32.Goffredi S., Childress J., Desaulniers N., Lee R., Lallier F., Hammond D., Inorganic carbon acquisition by the hydrothermal vent tubeworm Riftia pachyptila depends upon high external PCO2 and upon proton-equivalent ion transport by the worm. J. Exp. Biol. 200, 883–896 (1997). [DOI] [PubMed] [Google Scholar]
- 33.De Cian M. C., Bailly X., Morales J., Strub J. M., Van Dorsselaer A., Lallier F. H., Characterization of carbonic anhydrases from Riftia pachyptila, a symbiotic invertebrate from deep-sea hydrothermal vents. Proteins 51, 327–339 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Nyholm S. V., Song P., Dang J., Bunce C., Girguis P. R., Expression and putative function of innate immunity genes under in situ conditions in the symbiotic hydrothermal vent tubeworm Ridgeia piscesae. PLOS ONE 7, e38267 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nyholm S. V., Graf J., Knowing your friends: Invertebrate innate immunity fosters beneficial bacterial symbioses. Nat. Rev. Microbiol. 10, 815–827 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Y., Tassia M. G., Waits D. S., Bogantes V. E., David K. T., Halanych K. M., Genomic adaptations to chemosymbiosis in the deep-sea seep-dwelling tubeworm Lamellibrachia luymesi. BMC Biol. 17, 91 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uchida T., Yoshioka Y., Yoshida Y., Fujie M., Yamaki A., Sasaki A., Inoue K., Shinzato C., Genomic and transcriptomic analyses illuminate the molecular basis of the unique lifestyle of a tubeworm, Lamellibrachia satsuma. DNA Res. 30, dsad014 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun Y., Sun J., Yang Y., Lan Y., Ip J. C.-H., Wong W. C., Kwan Y. H., Zhang Y., Han Z., Qiu J.-W., Qian P. Y., Genomic signatures supporting the symbiosis and formation of chitinous tube in the deep-sea tubeworm Paraescarpia echinospica. Mol. Biol. Evol. 38, 4116–4134 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hinzke T., Kleiner M., Breusing C., Felbeck H., Hasler R., Sievert S. M., Schluter R., Rosenstiel P., Reusch T. B. H., Schweder T., Markert S., Host-microbe interactions in the chemosynthetic Riftia pachyptila symbiosis. mBio 10, e02243-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.García-Castro H., Kenny N. J., Iglesias M., Álvarez-Campos P., Mason V., Elek A., Schönauer A., Sleight V. A., Neiro J., Aboobaker A., Permanyer J., Irimia M., Sebé-Pedrós A., Solana J., ACME dissociation: A versatile cell fixation-dissociation method for single-cell transcriptomics. Genome Biol. 22, 89 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robinson J. J., Stein J. L., Cavanaugh C. M., Cloning and sequencing of a form II ribulose-1,5-biphosphate carboxylase/oxygenase from the bacterial symbiont of the hydrothermal vent tubeworm Riftia pachyptila. J. Bacteriol. 180, 1596–1599 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flores J. F., Fisher C. R., Carney S. L., Green B. N., Freytag J. K., Schaeffer S. W., Royer W. E. Jr., Sulfide binding is mediated by zinc ions discovered in the crystal structure of a hydrothermal vent tubeworm hemoglobin. Proc. Natl. Acad. Sci. U.S.A. 102, 2713–2718 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rimskaya-Korsakova N. N., Galkin S. V., Malakhov V. V., The anatomy of the blood vascular system of the giant vestimentiferan tubeworm Riftia pachyptila (Siboglinidae, Annelida). J. Morphol. 278, 810–827 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Wakabayashi T., Naito H., Cellular heterogeneity and stem cells of vascular endothelial cells in blood vessel formation and homeostasis: Insights from single-cell RNA sequencing. Front. Cell Dev. Biol. 11, 1146399 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang H., He K., Zhang H., Zhang Q., Cao L., Li J., Zhong Z., Chen H., Zhou L., Lian C., Wang M., Chen K., Qian P.-Y., Li C., Deciphering deep-sea chemosynthetic symbiosis by single-nucleus RNA-sequencing. eLife 12, RP88294 (2023). [Google Scholar]
- 46.Renoz F., Noel C., Errachid A., Foray V., Hance T., Infection dynamic of symbiotic bacteria in the pea aphid Acyrthosiphon pisum gut and host immune response at the early steps in the infection process. PLOS ONE 10, e0122099 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wikstrom M., Krab K., Sharma V., Oxygen activation and energy conservation by cytochrome c oxidase. Chem. Rev. 118, 2469–2490 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bottiglieri T., S-Adenosyl-L-methionine (SAMe): From the bench to the bedside—Molecular basis of a pleiotrophic molecule. Am. J. Clin. Nutr. 76, 1151S–1157S (2002). [DOI] [PubMed] [Google Scholar]
- 49.Struck A. W., Thompson M. L., Wong L. S., Micklefield J., S-adenosyl-methionine-dependent methyltransferases: Highly versatile enzymes in biocatalysis, biosynthesis and other biotechnological applications. Chembiochem 13, 2642–2655 (2012). [DOI] [PubMed] [Google Scholar]
- 50.Alvarez-Carreno C., Alva V., Becerra A., Lazcano A., Structure, function and evolution of the hemerythrin-like domain superfamily. Protein Sci. 27, 848–860 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanchez S., Andersen A. C., Hourdez S., Lallier F. H., Identification, sequencing, and localization of a new carbonic anhydrase transcript from the hydrothermal vent tubeworm Riftia pachyptila. FEBS J. 274, 5311–5324 (2007). [DOI] [PubMed] [Google Scholar]
- 52.Yan G., Lan Y., Sun J., Xu T., Wei T., Qian P.-Y., Comparative transcriptomic analysis of in situ and onboard fixed deep-sea limpets reveals sample preparation-related differences. iScience 25, 104092 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cao L., Lian C., Zhang X., Zhang H., Wang H., Zhou L., Wang M., Chen H., Luan Z., Li C., In situ detection of the fine scale heterogeneity of active cold seep environment of the Formosa Ridge, the South China Sea. J. Mar. Syst. 218, 103530 (2021). [Google Scholar]
- 54.Renosto F., Martin R. L., Borrell J. L., Nelson D. C., Segel I. H., ATP sulfurylase from trophosome tissue of Riftia pachyptila (hydrothermal vent tube worm). Arch. Biochem. Biophys. 290, 66–78 (1991). [DOI] [PubMed] [Google Scholar]
- 55.Laue B. E., Nelson D. C., Sulfur-oxidizing symbionts have not co-evolved with their hydrothermal vent tube worm hosts: An RFLP analysis. Mol. Mar. Biol. Biotechnol. 6, 180–188 (1997). [PubMed] [Google Scholar]
- 56.Bohnke S., Perner M., Seeking active RubisCOs from the currently uncultured microbial majority colonizing deep-sea hydrothermal vent environments. ISME J. 13, 2475–2488 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Duperthuy M., Sjostrom A. E., Sabharwal D., Damghani F., Uhlin B. E., Wai S. N., Role of the Vibrio cholerae matrix protein Bap1 in cross-resistance to antimicrobial peptides. PLOS Pathog. 9, e1003620 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ekici S., Pawlik G., Lohmeyer E., Koch H.-G., Daldal F., Biogenesis of cbb3-type cytochrome c oxidase in Rhodobacter capsulatus. Biochim. Biophys. Acta Bioenerg. 1817, 898–910 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.N. Wallace, A. Zani, E. Abrams, Y. Sun, “The impact of oxygen on bacterial enteric pathogens” in Advances in Applied Microbiology, vol. 95, S. Sariaslani, G. M. Gadd, Eds. (Academic Press, 2016), chap. 4, pp. 179–204. [DOI] [PubMed] [Google Scholar]
- 60.Vigneron A., Charif D., Vincent-Monegat C., Vallier A., Gavory F., Wincker P., Heddi A., Host gene response to endosymbiont and pathogen in the cereal weevil Sitophilus oryzae. BMC Microbiol. 12 (Suppl. 1), S14 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Simonet P., Gaget K., Balmand S., Ribeiro Lopes M., Parisot N., Buhler K., Duport G., Vulsteke V., Febvay G., Heddi A., Charles H., Callaerts P., Calevro F., Bacteriocyte cell death in the pea aphid/Buchnera symbiotic system. Proc. Natl. Acad. Sci. U.S.A. 115, E1819–E1828 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Isabella V. M., Lapek J. D. Jr., Kennedy E. M., Clark V. L., Functional analysis of NsrR, a nitric oxide-sensing Rrf2 repressor in Neisseria gonorrhoeae. Mol. Microbiol. 71, 227–239 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fritz G., Roth A., Schiffer A., Buchert T., Bourenkov G., Bartunik H. D., Huber H., Stetter K. O., Kroneck P. M., Ermler U., Structure of adenylylsulfate reductase from the hyperthermophilic Archaeoglobus fulgidus at 1.6-A resolution. Proc. Natl. Acad. Sci. U.S.A. 99, 1836–1841 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fu Y., Tang J., Yao G. F., Huang Z. Q., Li Y. H., Han Z., Chen X. Y., Hu L. Y., Hu K. D., Zhang H., Central role of adenosine 5′-phosphosulfate reductase in the control of plant hydrogen sulfide metabolism. Front. Plant Sci. 9, 1404 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Duperthuy M., Schmitt P., Garzon E., Caro A., Rosa R. D., Le Roux F., Lautredou-Audouy N., Got P., Romestand B., de Lorgeril J., Kieffer-Jaquinod S., Bachere E., Destoumieux-Garzon D., Use of OmpU porins for attachment and invasion of Crassostrea gigas immune cells by the oyster pathogen Vibrio splendidus. Proc. Natl. Acad. Sci. U.S.A. 108, 2993–2998 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu W., Huang L., Mao Q., Wei J., Li J., Zhao Y., Zhang Q., Jia D., Wei T., Interaction of viral pathogen with porin channels on the outer membrane of insect bacterial symbionts mediates their joint transovarial transmission. Philos. Trans. R. Soc. Lond. B Biol. Sci. 374, 20180320 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang H., Liu Z., Ma S., Zhang H., Kong F., He Y., Yang X., Wang Y., Xu H., Yang A., Tian J., Zhang M., Cao J., Jiang Y., Guo X., Ratio of S-adenosylmethionine to S-adenosylhomocysteine as a sensitive indicator of atherosclerosis. Mol. Med. Rep. 14, 289–300 (2016). [DOI] [PubMed] [Google Scholar]
- 68.Sbodio J. I., Snyder S. H., Paul B. D., Regulators of the transsulfuration pathway. Br. J. Pharmacol. 176, 583–593 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.D. Weihrauch, M. O'Donnell, Acid-Base Balance and Nitrogen Excretion in Invertebrates: Mechanisms and Strategies in Various Invertebrate Groups with Considerations of Challenges Caused by Ocean Acidification (Springer, 2017), 306 pp. [Google Scholar]
- 70.Enroth C., Eger B. T., Okamoto K., Nishino T., Nishino T., Pai E. F., Crystal structures of bovine milk xanthine dehydrogenase and xanthine oxidase: Structure-based mechanism of conversion. Proc. Natl. Acad. Sci. U.S.A. 97, 10723–10728 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Larsen E. H., Deaton L. E., Onken H., O'Donnell M., Grosell M., Dantzler W. H., Weihrauch D., Osmoregulation and excretion. Compr. Physiol. 4, 405–573 (2014). [DOI] [PubMed] [Google Scholar]
- 72.Campbell E. M., Ball A., Hoppler S., Bowman A. S., Invertebrate aquaporins: A review. J. Comp. Physiol. B 178, 935–955 (2008). [DOI] [PubMed] [Google Scholar]
- 73.Litman T., Sogaard R., Zeuthen T., Ammonia and urea permeability of mammalian aquaporins. Handb. Exp. Pharmacol. 190, 327–358 (2009). [DOI] [PubMed] [Google Scholar]
- 74.Wang M. F. Z., Mantri M., Chou S.-P., Scuderi G. J., McKellar D. W., Butcher J. T., Danko C. G., De Vlaminck I., Uncovering transcriptional dark matter via gene annotation independent single-cell RNA sequencing analysis. Nat. Commun. 12, 2158 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sun Y., Liang Q., Sun J., Yang Y., Tao J., Liang J., Feng D., Qiu J.-W., Qian P.-Y., The mitochondrial genome of the deep-sea tubeworm Paraescarpia echinospica (Siboglinidae, Annelida) and its phylogenetic implications. Mitochondrial DNA B Resour. 3, 131–132 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hu M., Zheng X., Fan C. M., Zheng Y., Lineage dynamics of the endosymbiotic cell type in the soft coral Xenia. Nature 582, 534–538 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sebé-Pedrós A., Saudemont B., Chomsky E., Plessier F., Mailhé M.-P., Renno J., Loe-Mie Y., Lifshitz A., Mukamel Z., Schmutz S., Novault S., Steinmetz P. R. H., Spitz F., Tanay A., Marlow H., Cnidarian cell type diversity and regulation revealed by whole-organism single-cell RNA-Seq. Cell 173, 1520–1534.e20 (2018). [DOI] [PubMed] [Google Scholar]
- 78.Hao Y., Hao S., Andersen-Nissen E., Mauck W. M. III, Zheng S., Butler A., Lee M. J., Wilk A. J., Darby C., Zager M., Hoffman P., Stoeckius M., Papalexi E., Mimitou E. P., Jain J., Srivastava A., Stuart T., Fleming L. M., Yeung B., Rogers A. J., McElrath J. M., Blish C. A., Gottardo R., Smibert P., Satija R., Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., Feng T., Zhou L., Tang W., Zhan L., Fu X., Liu S., Bo X., Yu G., clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (Camb) 2, 100141 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cao J., Spielmann M., Qiu X., Huang X., Ibrahim D. M., Hill A. J., Zhang F., Mundlos S., Christiansen L., Steemers F. J., Trapnell C., Shendure J., The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N. J., Livak K. J., Mikkelsen T. S., Rinn J. L., The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Amann R. I., Binder B. J., Olson R. J., Chisholm S. W., Devereux R., Stahl D. A., Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56, 1919–1925 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Thisse C., Thisse B., High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59–69 (2008). [DOI] [PubMed] [Google Scholar]
- 84.Choi H. M. T., Schwarzkopf M., Fornace M. E., Acharya A., Artavanis G., Stegmaier J., Cunha A., Pierce N. A., Third-generation in situ hybridization chain reaction: Multiplexed, quantitative, sensitive, versatile, robust. Development 145, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S20
Legends for tables S1 to S6
Tables S1 to S6