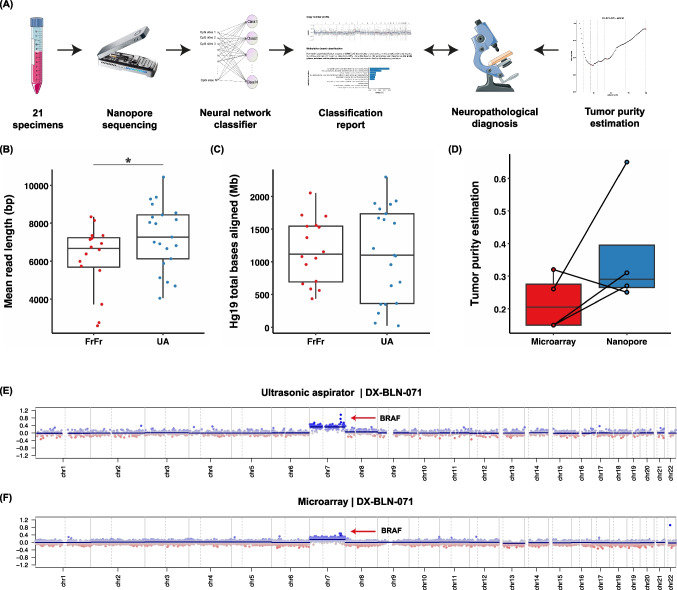
Fig. 1.
(A) Overview of the study design including workflow using ultrasonic aspirator tissue specimens for nanopore sequencing for DNA methylation-based classification using a neural network classifier and copy number variation analysis, comparison to microarray-based routine neuropathological profiling and assessment of tumor purity by absolute copy number estimation using ACE. Suitability of ultrasonic aspirator-derived tumor tissue (UA) for nanopore sequencing: (B,C) Comparison of (B) mean read length and (C) read yield obtained from standard nanopore protocol using fresh-frozen (FrFr) tumor tissue vs. ultrasonic aspirator-derived sample material indicates similar sequencing performance. Asterisk indicates p-value < 0.05 for group comparison by Student’s t-test. (D) In silico tumor purity estimation between nanopore ultrasonic aspirator tissue samples and microarray FFPE tissue. (E,F) Representative illustration of matched copy number variation profiles obtained from (E) ultrasonic aspirator tissue samples and nanopore sequencing and (F) FFPE tumor tissue subjected to EPIC microarray (850 K). Red marker indicates a low-level gain of the BRAF locus suggestive of a BRAF gene fusion