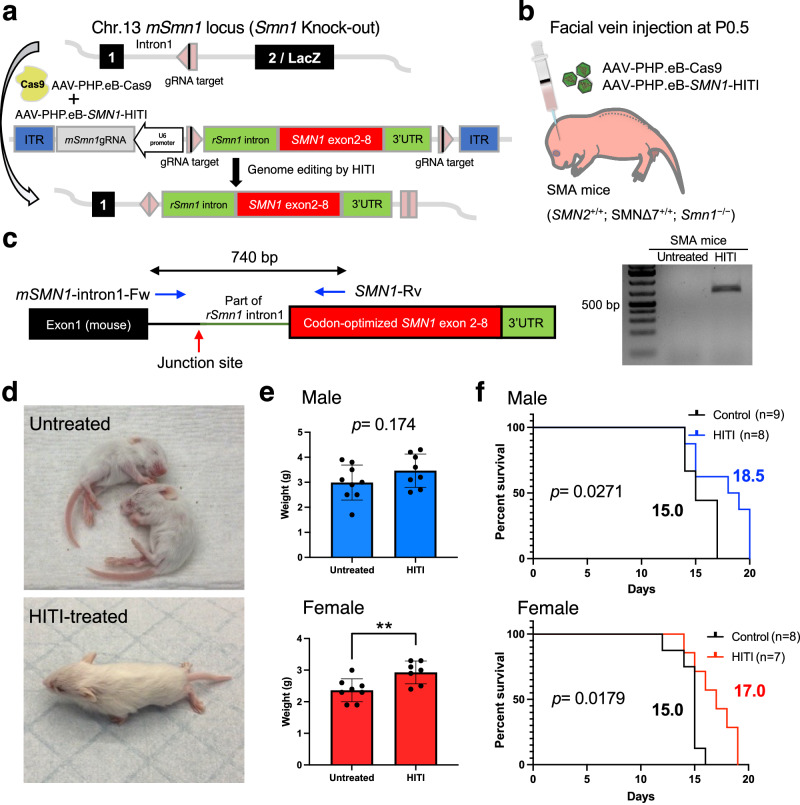
Fig. 2. HITI-mediated gene correction of SMA mice.
a Schematic of gene correction in SMA mice (SMN2+/+; SMNΔ7+/+; Smn1−/−). gRNA is expressed under the U6 promoter. Rat Smn1 intron, optimized mouse exon 2–8 and rat 3’UTR is sandwiched by Cas9/gRNA target sequence. The Cas9/gRNA creates a DSB at target sites and releases the donor. The donor sequence is integrated into the target site by NHEJ-based repair. Pink pentagon, Cas9/gRNA target sequence. Black line within the pink pentagon, Cas9 cleavage site. b AAVs (1 × 1011GC of each AAV) were systemically delivered via the facial vein in neonatal SMA mice. Body weight and life span were recorded after the injection. c The region between mouse intron1 and inserted rat intron1-optimized exon can be detected only in HITI-treated SMA sample by PCR. Blue arrows denote the location of the primers. Red arrow denotes the location of the junction site. d Gross morphology of untreated SMA mice (upper) and HITI-treated SMA mouse (lower) at 2 weeks old. e Body weight comparison between untreated and HITI-treated SMA mice of 12 days old with the males depicted on the top (Untreated, n = 9; HITI-treated, n = 8), and the females depicted on the bottom (Untreated, n = 8; HITI-treated, n = 7). Data are represented as mean ± S.D. **p = 0.0096, a two-sided unpaired Student’s t test. f Survival rate comparison between untreated and HITI-treated SMA mice with the males depicted on the top, and the females depicted on the bottom. n is the number of animals per group. Log-rank (Mantel–Cox) test was used for survival curves. The p value and median survival are indicated for all curves. Source data are provided as a Source Data file.