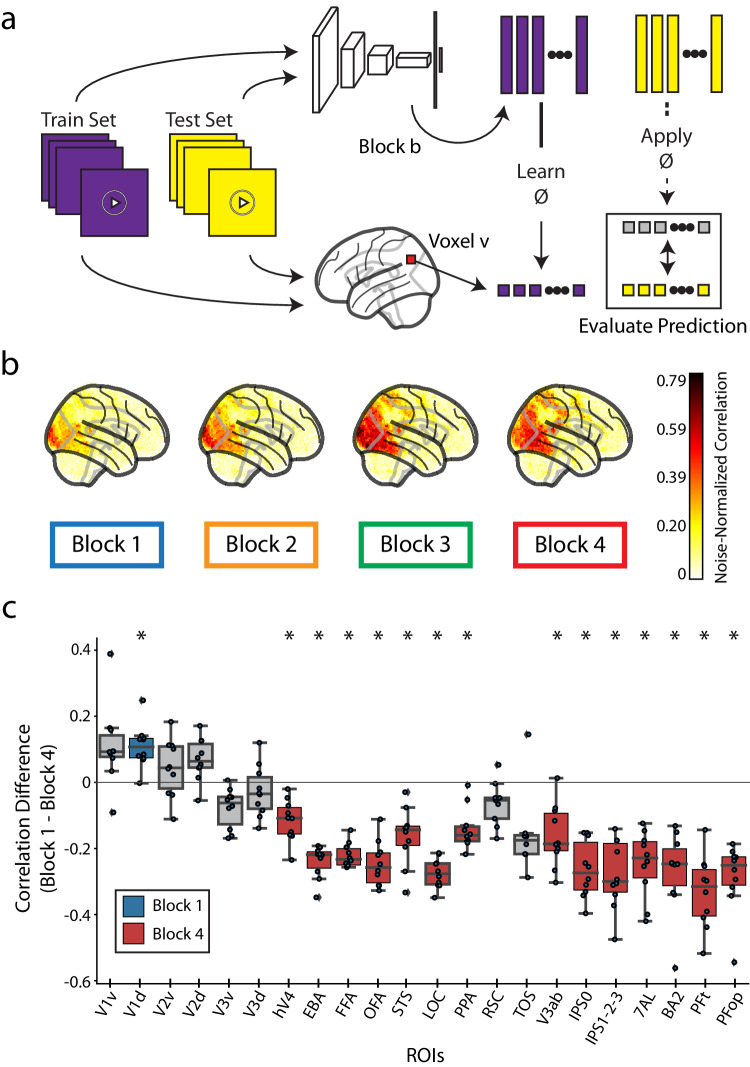
Fig. 4. Evaluation of biologically-similar video-based encoding model.
a Voxelwise encoding model procedure: All videos are shown to both a TSM ResNet50 DNN and a human. Training set video embeddings are extracted from a block b of the DNN and used to learn a voxelwise mapping function to the human responses. This mapping is then applied to the testing set video embeddings to predict the brain response at each voxel. b Whole-brain encoding accuracy across blocks: We use the encoding model procedure with each of the four blocks of a TSM ResNet50 model trained to recognize actions in videos to predict the neural response at each voxel in the whole brain. The brain figures show the subject-average noise-normalized predictive correlation (divided by the voxel’s upper noise ceiling) at each voxel. c ROI-based encoding accuracy difference: Difference in predictive performance between block 1 and block 4 at each of the 22 ROIs. Predictive performance at each voxel is measured as the noise-normalized correlation between the brain responses and the predicted responses, averaged over all reliable voxels in each ROI. Significant ROIs are denoted with an asterisk and a color (blue for Block 1, red for Block 4, gray is not significant) corresponding to the significant layer (p < 0.05, one sample two-sided t-test against a population mean of 0, Bonferroni corrected across n = 22 ROIs). Source data are provided as a Source Data file. The box plot encompasses the first and third data quartiles and the median (horizontal line). The whiskers extend to the minimum and maximum values within 1.5 times the interquartile range, and values falling outside that range are considered outliers (denoted by a diamond). The overlaid points show the value at each observation (n = 10 for all ROIs except transverse occipital sulcus (TOS, n = 8) and retrosplenial cortex (RSC, n = 9)).