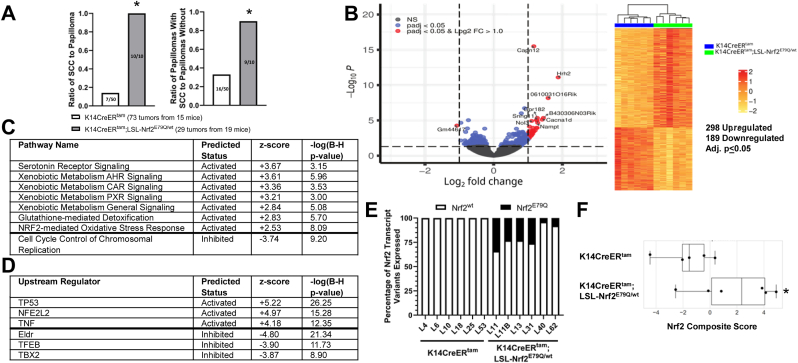
Fig. 5.
Remaining Nrf2E79Q activated tumors display enhanced progression and enrichment of Nrf2 target gene expression. (A) Ratio of SCC to papilloma and ratio of papilloma with SCC to papilloma from tumors collected from 15 K14CreERtam and 19 K14CreERtam;LSL-Nrf2E79Q/wt mice as determined by H&E histological analysis. * Denotes p ≤ 0.05 Fisher's Exact Test and p ≤ 0.05 Risk Difference. (B) Volcano plot and heatmap of tumor RNAseq data from K14CreERtam;LSL-Nrf2E79Q/wt skin tumors compared to K14CreERtam skin tumors. Each column represents a tumor from a different mouse. (C) IPA pathway analysis of RNAseq data reveals, of the significantly enriched pathways, the top pathways predicted to be activated (z-score ≥2.0) and pathways predicted to be inhibited (z-score ≤2.0) in K14CreERtam;LSL-Nrf2E79Q/wt skin tumors compared to K14CreERtam skin tumors. Right tailed Fisher Exact Test with Benjamini-Hochberg (B–H) multiple hypothesis testing-corrected p-value (D) IPA's upstream regulator analysis of RNAseq data reveals top upstream transcription regulators, upstream regulators with z-scores ≥2.0 are predicted to be activated and upstream regulators with z-scores ≤2.0 are predicted to be inhibited based on observed gene expression changes in tumors of K14CreERtam;LSL-Nrf2E79Q/wt mice compared to K14CreERtam mice. Right tailed Fisher Exact Test with Benjamini-Hochberg (B–H) multiple hypothesis testing-corrected p-value (E) Tumors from K14CreERtam;LSL-Nrf2E79Q/wt epidermis expresses Nrf2E79Q transcripts. Each bar represents a tumor from a different mouse. (F) OIS-PRM targeted proteomics showed an increase in the Nrf2 score in tumors from K14CreERtam;LSL-Nrf2E79Q/wt mice compared to tumors from K14CreERtam mice. * Denotes p = 0.08 via Mann-Whitney U test.