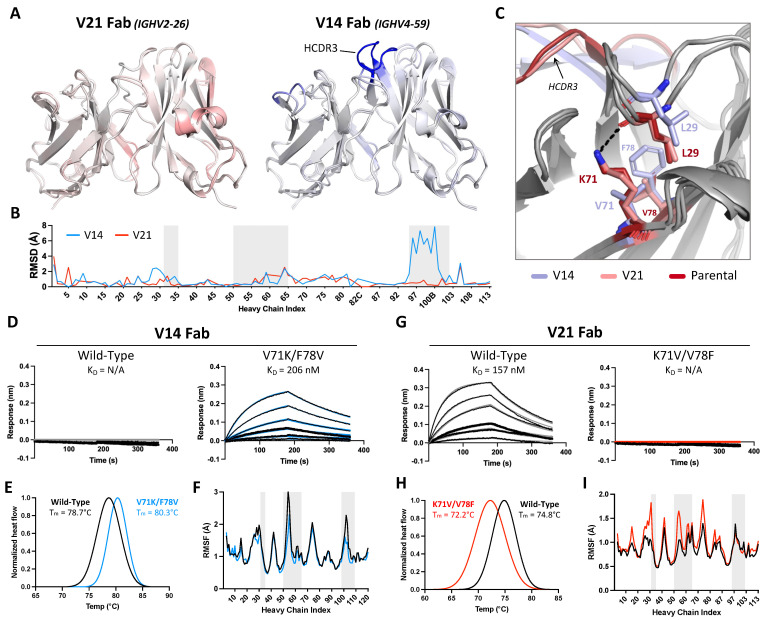
Figure 3.
Framework residues in the 44H10 heavy chain modulate antibody paratope stability and antigen binding. (A) Overlay of V21 (left) and V14 (right) Fab variable region crystal structures with parental 44H10 colored by RMSD, where increasing saturation indicates greater deviation from the parental structure. (B) Quantification of V14 and V21 heavy chain variable regions RMSDs relative to parental 44H10. Areas with gray shading indicate Kabat HCDRs. (C) Close-up view of stabilizing interactions mediated by residues H-K71 and H-V78 with H-L29 in parental 44H10 (red) and V21 (salmon), which cannot be mediated by H-V71 and H-F78 in V14 (light blue). A black dotted line indicates a hydrogen bond. (D–I) Biophysical characterization of wild-type V14 Fab and mutant V14 Fab with V71K/F78V heavy chain mutations (D–F) and wild-type V21 Fab and mutant V21 Fab with K71V/V78F heavy chain mutations (G–I). (D,G) BLI binding profiles of V14 and V21 wild-type and mutant Fabs to recombinant HLA-DR, where black lines represent measured binding and colored curves correspond to the data fitted to a 1:1 binding model. (E,H) Differential Scanning Calorimetry (DSC) profiles of V14 and V21 wild-type and mutant Fabs with calculated melting temperatures (Tm). (F,I) RMSF plots derived from MD simulations of V14 and V21 wild-type and mutant Fabs indicating changes in heavy chain variable region flexibility. Areas with gray shading represent Kabat HCDRs.