Fig. 4.
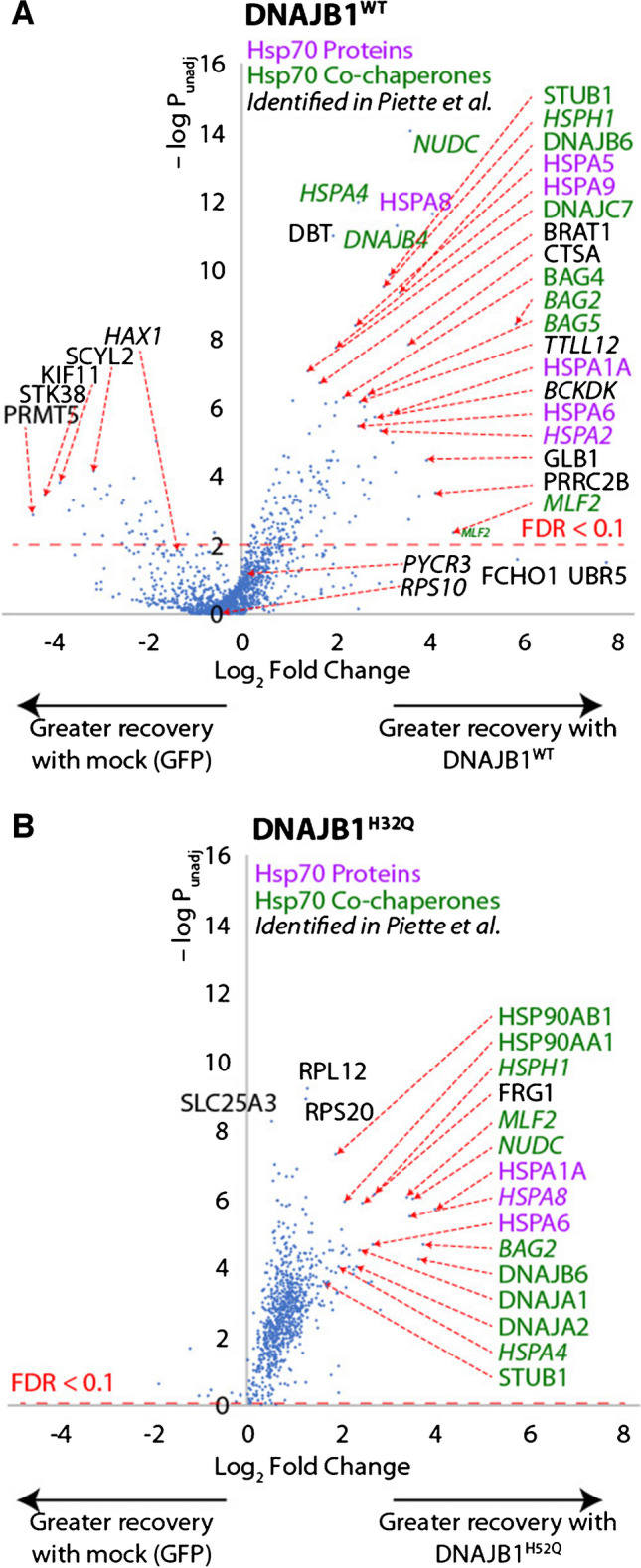
A Proteins recovered by immunoprecipitation of FlagDNAJB1WT without crosslinking (comparable to Fig. 2B) and B proteins recovered by immunoprecipitation of FlagDNAJB1H32Q with crosslinking (comparable to ref. 8). Mock cells are transfected with eGFP. The experimental strategy is similar to that from Fig. 2A, except that the DNAJB1H32Q AP-MS was preceded by cellular crosslinking with 250 µM DSP. Nine biological replicates, comprising three LC–MS runs, were performed for DNAJB1WT, while six biological replicates, comprising two LC–MS runs, were performed for DNAJB1H32Q. Hsp70 proteins are indicated in purple, Hsp70-associated co-chaperones are in green, and protein identified from Piette et al. [31] are italicized. The p-value cut-off corresponding to a 10% false discovery rate is indicated. The global false discovery rate for all proteins from Storey’s modification of Benjamini–Hochberg analysis is 73% for DNAJB1WT and 5.9% for DNAJB1H32Q
