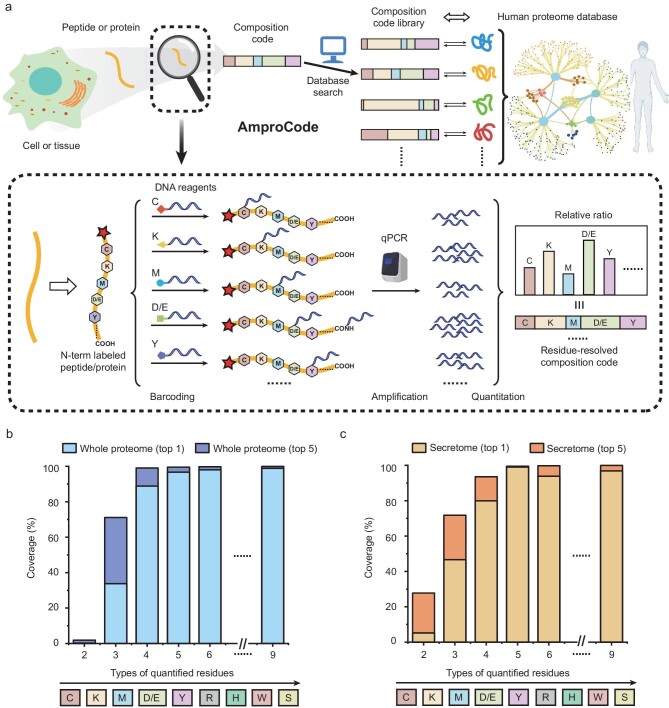
Figure 1.
Design and overview of AmproCode. (a) The workflow of amplifiable protein identification through AmproCode. Each type of selected residue on peptide or protein samples is modified by using DNA barcodes, respectively. The residue-resolved composition code is generated by the relative ratio of these residues from each protein, which can be amplified and quantified by using qPCR. The sequence information of all proteins in the human proteome database can be converted into the composition code library. Matching the experimentally obtained composition code with the codes in the database library by using the database search algorithm allows the identification of the sample. (b) Theoretical evaluation of the coverage rate of AmproCode in the whole human proteome. (c) Theoretical evaluation of the coverage rate of AmproCode in the secretome. The residue types in the composition code range from two to nine in the order of Cys, Lys, Met, Asp/Glu, Tyr, Arg, His, Trp and Ser. Targeting the highest single hit and the top five hits in identification are evaluated respectively.