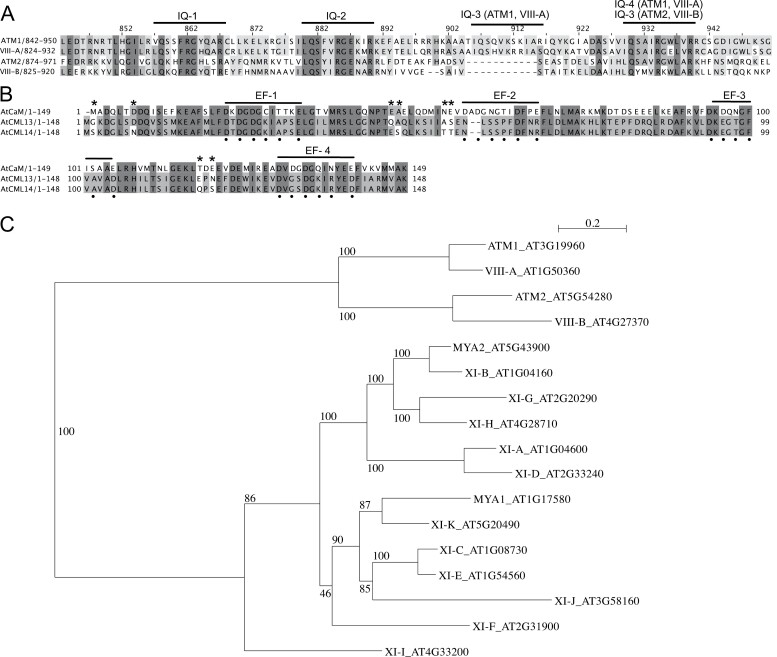
Fig. 1.
Protein sequence alignments of (A) the neck regions (residues ~800–1000) for Arabidopsis class VIII myosins, and (B) Arabidopsis CaM (AtCaM7, AT3G43810), CML13, and CML14. Amino acid residues were shaded based on their percentage identity, dark gray if identical, and progressively lighter gray through to white as unconserved. In (A), predicted IQ motif consensus sequences are overlined. The EF-hands of CaM in (B) are overlined, and Ca2+-coordinating residues in CaM are marked with a dot beneath the alignment. Residues that differ between CML13 and CML14 are indicated with an asterisk. ClustalΩ was used for alignment (Sievers and Higgins, 2014; Gouy et al., 2021), and images were generated using Jalview Version 2.11.2.6. (C) Phylogenetic tree of Arabidopsis class VIII and XI myosins. The tree was constructed using Seaview version 5.0 (Sievers and Higgins, 2014; Gouy et al., 2021) from a complete alignment of proteins using PhyML with bootstrapping analysis (1000 replicates).