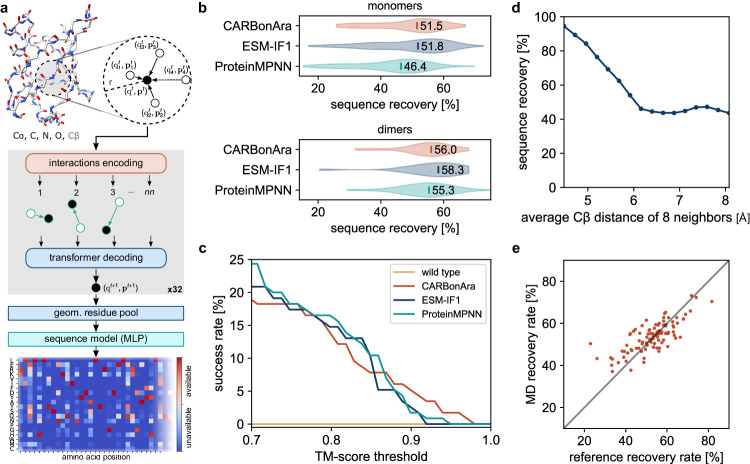
Fig. 1. CARBonAra’s architecture and comparison with state-of-the-art methods.
a The model applies multiple geometric transformer operations to the coordinates and atom element of a backbone scaffold with added virtual Cβ to predict the amino acid confidence at each position in the sequence expressed as a position-specific scoring matrix. b Comparison of the sequence recovery of different methods for monomers and dimers with indicated median sequence recovery. c Percentage of AlphaFold predicted structures, in single-sequence mode, above a TM-score threshold using sequences sampled in the same way for different methods. d Sequence recovery as a function of the average Cβ distance of the 8 nearest neighbors. e Consensus prediction recovery rate against the reference experimental structure recovery rate, derived from 500 frames sampled from 1 μs molecular dynamics simulations for 80 monomers.