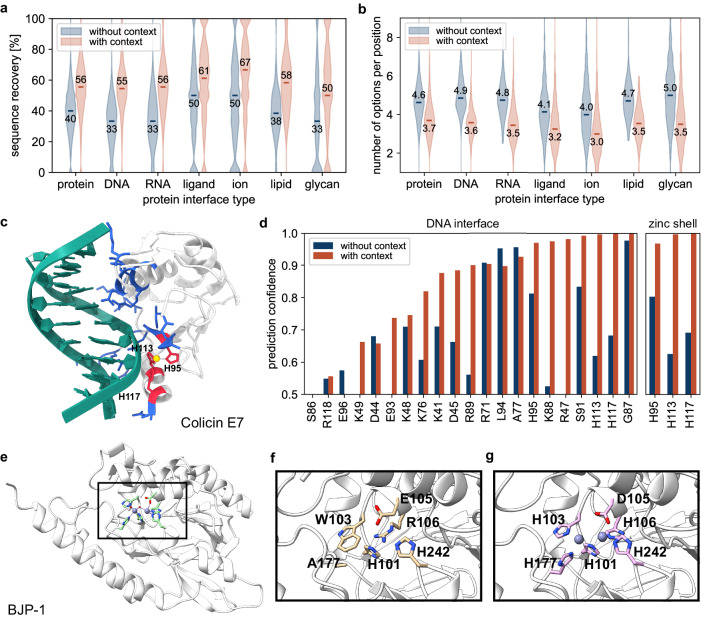
Fig. 3. Context-aware amino acid recovery allows the design of functional proteins.
a Sequence recovery at the interface (residues within 5 Å) without and with proteins, nucleic acids, ligands, ions, lipids, and glycans binders. b Number of predicted possible amino acids per position at the interface (residues within 5 Å) without and with proteins, nucleic acids, ligands, ions, lipids, and glycans binders (considering a confidence prediction threshold of 0.5). c Colicin E7 endonuclease domain in complex with DNA and a zinc ion (PDB ID: 1ZNS). The protein-DNA interface (residues within 4 Å) is highlighted in blue. The protein-zinc shell is highlighted in red (residues within 3 Å). d Estimated accurate prediction probability for the scaffold amino acids at the protein-DNA interface and the protein-zinc shell with and without the presence of DNA and zinc. e Metallo β-lactamase structure of BJP−1 with the catalytic pocket containing two zinc ions (PDB ID: 3LVZ). The pocket of an AlphaFold predicted structure from a CARBonAra-designed sequence applied to the scaffold backbone without zinc ions (f) and containing the original zinc ions (g).