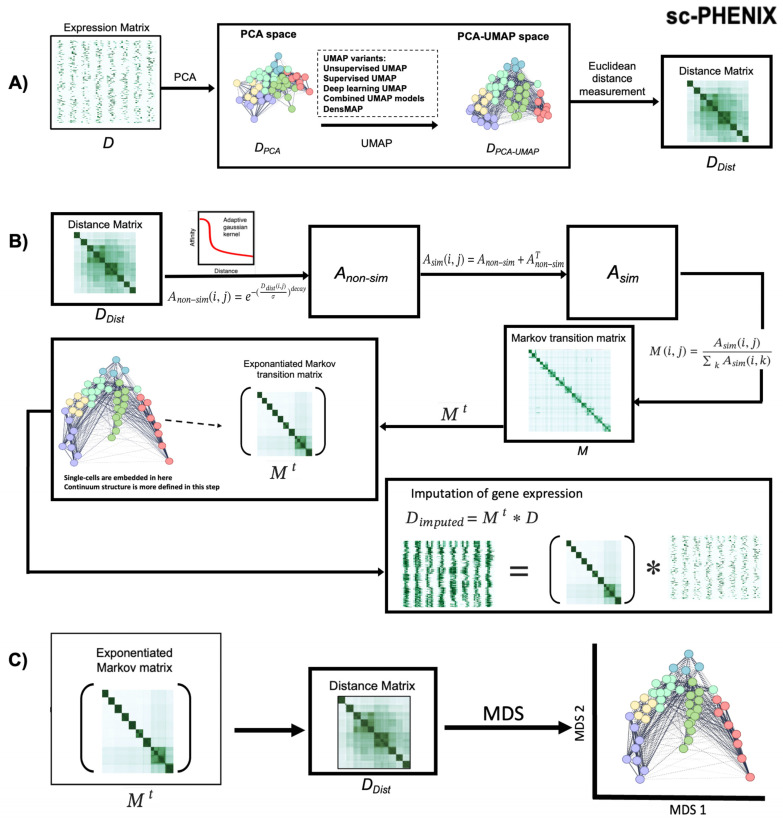
Figure 1.
The imputation process using sc-PHENIX. The sc-PHENIX imputation approach for scRNA-seq data consists of two main steps: (A) The construction of the distance matrix (): sc-PHENIX is characterized by applying PCA and then UMAP (PCA-UMAP). In this PCA-UMAP multidimensional space, sc-PHENIX constructs the best denoise representation of cell distance measurements for the diffusion process to preserve data structures. (B) The diffusion maps for imputation: the imputation process using diffusion maps consists of several steps: (i) Construction of the Markov transition matrix M from : sc-PHENIX uses the adaptive Gaussian kernel to generate a non-symmetric affinity matrix (), it is symmetrized. Then, it is normalized to generate (M). (ii) Diffusion process: M is exponentiated to a chosen power t (random walk of length t named “diffusion time”) to obtain the exponentiated Markov matrix (Mt). The Mt graph well preserves the continuum structure better than the previous steps. (iii) Imputation: This step consists of multiplying the exponentiated Markov matrix (Mt) times the single-cell-matrix data D to obtain an imputed and denoised scRNA-seq matrix (). Note: The symbol * used in this figure indicates matrix multiplication for Mt and D in a computational formalism, which is equivalent to the formal mathematical notation Mt ⋅ D. All equations are described in the Section 2 section. (C) Visualization of the exponentiated Markov matrix: We convert the Mt into a distance matrix (). Then, we apply a multidimensional scaling method to project data in 2D or 3D dimensions. This projection can be used as a heuristic method for quality control of imputation.