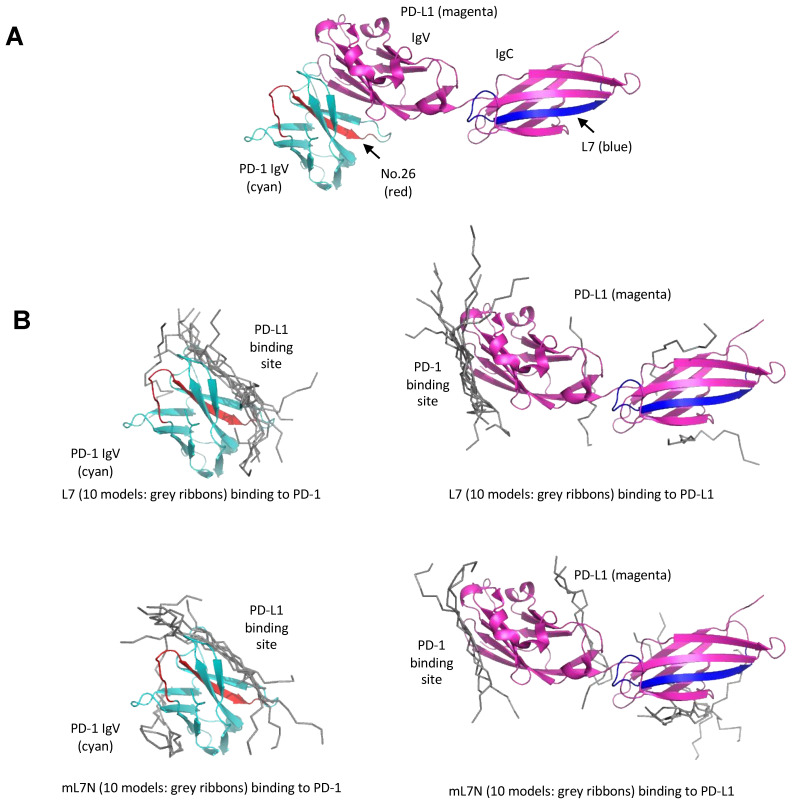
Figure 5.
Models for binding of the peptide blockers to the PD-1/PD-L1 ectodomains. (A) Peptide #26 is located at the interface between the PD-1 and PD-L1 IgV domains. The structure shown is a composite of the complex structure between the PD-1 and PD-L1 IgV domains (PDB code: 4ZQK). The PD-L1 structure shown contains both the IgV and IgC domains (PDB code: 4Z18). PD-1 is in cyan, while PD-L1 is in magenta. The region corresponding to the PD-1-derived peptide #26 (PD-1 Ser57-Ser71) is shown in orange red. The region corresponding to PD-L1-derived L7 peptide (PD-L1 Arg186-Thr201) is shown in blue. (B) Docking of the L7 or mL7N peptide to either the PD-1 or PD-L1 ectodomain. The HPEPDOCK server was used for prediction. The top 10 predicted peptide models from each complex calculation are shown in grey ribbons. The L7 and mL7N peptides were predicted to bind similarly to the PD-1 or PD-L1 IgV domains, which resemble each other in structure. The predicted binding site for L7/mL7N is the interface between PD-1 and PD-L1, as shown in (A).