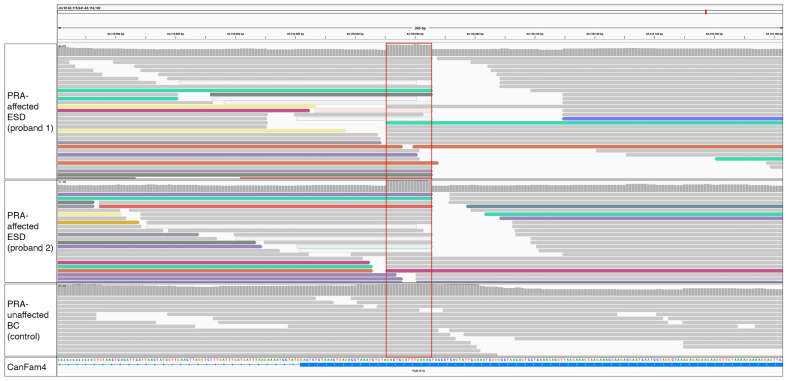
Figure 2.
WGS reads over a 243 bp region of FAM161A inclusive of exon 4 of probands 1 and 2 and a control PRA-unaffected Border Collie dog. WGS reads are based on the canine reference genome UU_Cfam_GSD_1.0 (CanFam4). Alignments represent the two PRA-affected probands and a PRA-unaffected Border Collie that acted as a control. Alignments were coloured by ‘Insert-Size and Pair Orientation’ and reads surrounding the location of increased read depth varied in colour, indicating that read mates aligned to different chromosomes. The red-boxed area (chr10: 63,116,051 to chr10: 63,116,065) is located within exon 4 of FAM161A within the reference genome. The increased read depth is indicative of a duplication or insertion, absent in the control WGS. Grey reads are aligning to the reference genome and coloured reads indicate reads aligning to alternate chromosomes typical of a repetitive element insertion.