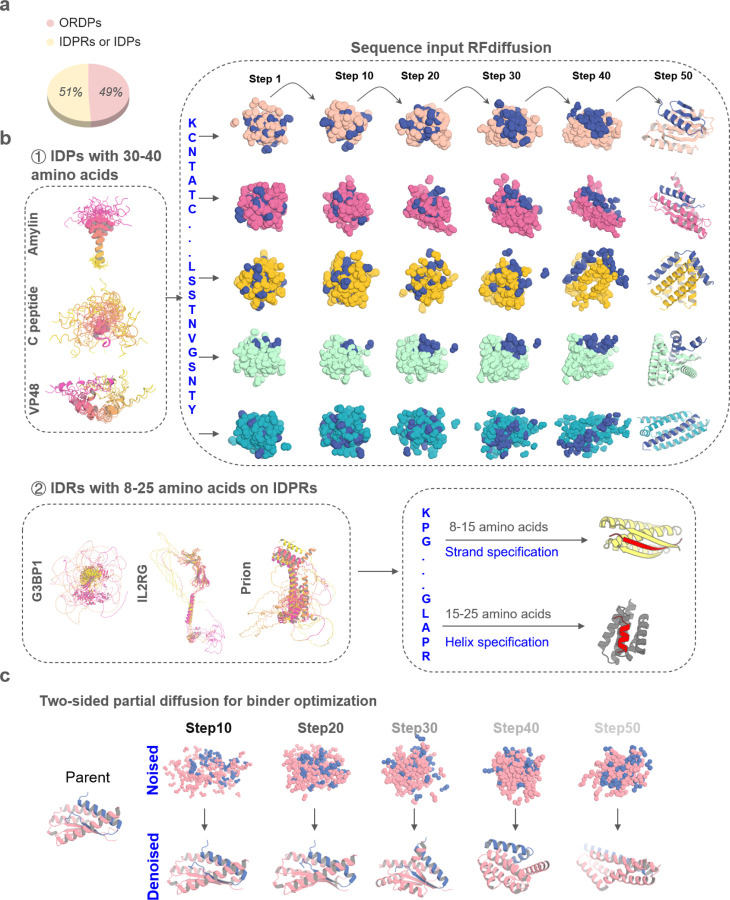
Figure. 1. Design strategies for binding conformational flexible peptides.
a, Frequency of ORDPs (ordered proteins), IDPRs /IDPs (intrinsically disordered proteins) in the human proteome41. b, ① Left, the NMR structure of Amylin (PDBID: 2KB8), C peptide (PDBID: 1T0C), the predicted structures of VP48 by five AlphaFold models22. The 5 predicted structures of VP48 are aligned together, revealing the flexibility of the intrinsically disordered protein. Right, Diffusion models for proteins are trained to recover noised protein structures and to generate new structures by reversing the corruption process through iterative denoising of initially random noise into a realistic structure. Here, A modified version of RFdiffusion was trained on two chain systems from the PDB to permit the design of protein binders to targets, for which only the sequence of the target was specified. The fine-tuned was found to generate binders to peptides in finely varying helix conformationsWith solely sequence input. ② Left, the predicted structures of G3BP1, IL2RG and prion by five AlphaFold models22. Right, A modified version of RFdiffusion was trained, allowing for specification of the secondary structure of a region, along with its sequence (See Method). When provided with the same target sequence input but different secondary structure specifications (helix or strand), the resulting conformations of the target could vary. c, Top: two sided partial diffusion. RFdiffusion is used to denoise a randomly noised starting parent design for both target and binder ; varying the extent by different noised step of initial noising (top row) enables control over the extent of introduced structural variation (bottom row; colours, new designs; grey, parent design).