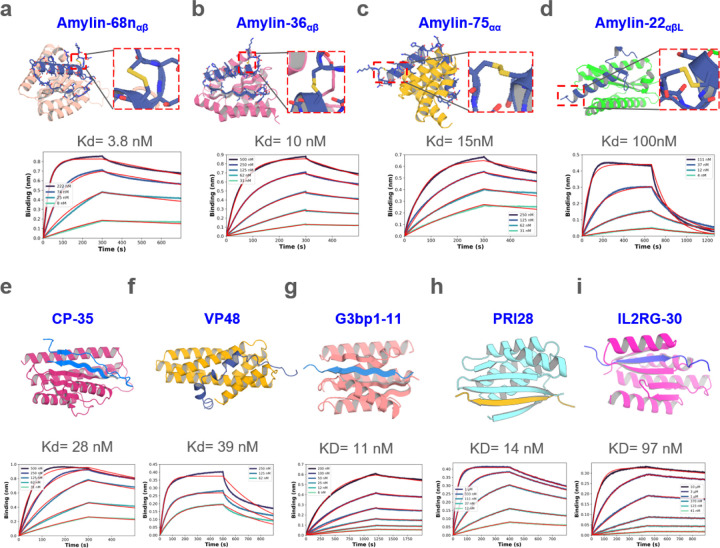
Figure. 2. Design of disordered region binder.
a-d, Binder design of Amylin using sequence input diffusion. Top, from left to right, design model of Amylin and its binder Amylin-68nαβ, Amylin-36αβ, Amylin-75αα and Amylin-22αβL, respectively. The secondary structure of Amylin is indicated in the subscript of the binder’s name. For each of the designs, the Amylin disulfide bonds between 2nd Cysteine and 7th Cysteine were retained well. Bottom, from left to right, the BLI measurement indicated that the binding affinity between Amylin-68nαβ, Amylin-36αβ, Amylin-75αα, Amylin-22αβL and Amylin are 3.8, 10, 15, 100 nM respectively. e-f, Binder design of CP and VP48 using sequence input diffusion, the binder affinity of CP and VP48 are 28 and 39 nM, respectively. g-i, Binder design using strand specification. Top, from left to right, design model of G3BP1RBD, prion and IL2RG and their binders G3bp1–11, PRI28 and IL2RG-30. Bottom, the BLI measurement indicated that the binding affinity of G3bp1–11, PRI28 and IL2RG-30 binders are 11, 14 and 97 nM, respectively.