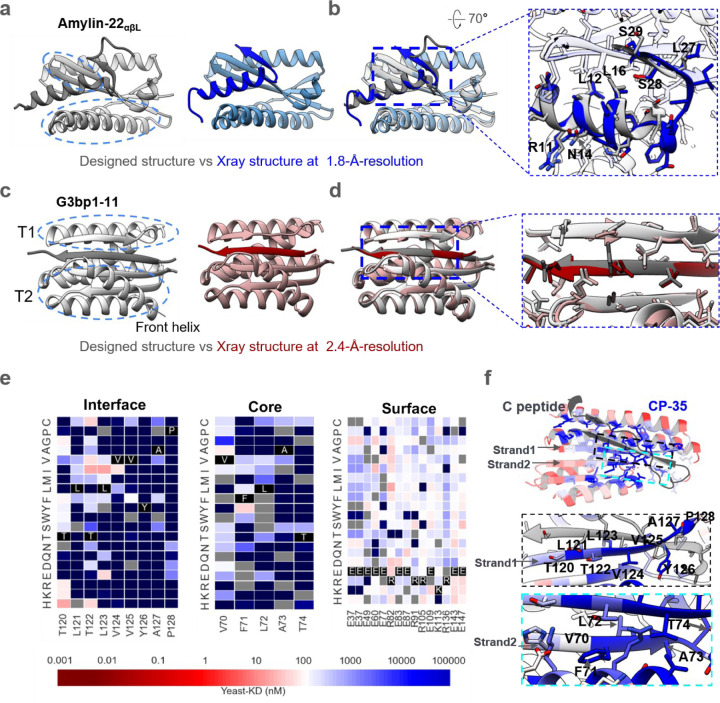
Figure. 3. Structural characterizations.
a, Left, the designed model of Amylin-22αβL, with target and binder proteins rendered in dim gray and gray, respectively. The helical and strand segments that create the groove in the binder, docking the helical segment of Amylin, are highlighted with blue dashed ellipsoid. Right, the crystal structure of Amylin-22αβL at 1.8 Å-resolution, with target and binder proteins rendered in blue and cornflower blue, respectively. b, Left, the overlay of the design model and the crystal structure of Amylin-22αβL. Right, magnified views of the regions indicated with black dotted frames in the left panel are provided to illustrate the detailed interface view of the design and crystal structure. The binder proteins are rendered with 90% transparency to enhance the visibility of the peptide target. The key residues on the Amylin are labeled to illustrate the good alignment of the key residues between designed protein and crystal structure. c, Left, the designed model of G3bp1–11, with target and binder proteins rendered in dim gray and gray, respectively. The two α/β topologies (T1 and T2) of the binders, forming the cleft where the target strand is positioned, are highlighted with blue dashed ellipses. The front helix of T2 is denoted by a black arrow. Right, the crystal structure of G3bp1–11 at 2.4 Å-resolution, with target and binder proteins rendered in dark red and rosy brown, respectively. d, Left, the overlay of the design model and the crystal structure of G3bp1–11. Right, magnified views of the regions indicated with black dotted frames in the left panel. The front helix of T2 has been surface capped to reveal the strand pairing interface. e, Heat maps representing C peptide-binding Kd (nM) values for single mutations in the designed interface (left), core (middle) and the surface (right). Substitutions that are heavily depleted are shown in blue, and beneficial mutations are shown in red, gray color indicates the lost yeast strains. For the interface region, we highlighted and showcased strand 1 (indicated by the arrow), which serves as the primary interaction secondary structure with the C peptide. For the core region, we showcased the right segment of strand 2 (indicated by the arrow), representing a main core region that does not form interactions with the C peptide. For the surface region, we selected the most exposed surface residues that don’t form any connections with other residues (Supplementary Fig. 6c). Full SSM map over all positions for CP35 is provided in Supplementary Fig. 6b. f, Top, designed binding proteins are colored by positional Shannon entropy from site saturation mutagenesis, with blue indicating positions of low entropy (conserved) and red those of high entropy (not conserved). Bottom, zoomed-in views of central regions of the design interface and core with the C peptide.