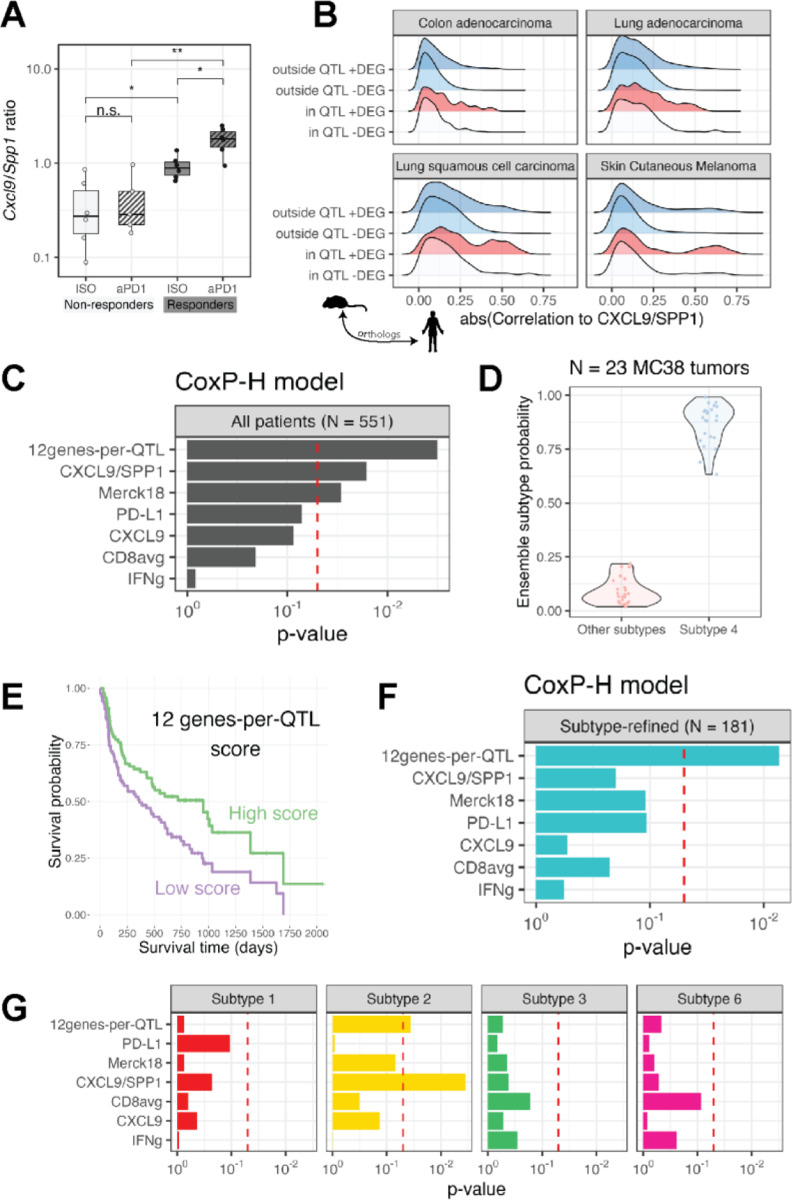
Figure 4: ST algorithm prioritizes genes within murine QTL predicting survival in specific human cancer immune subtypes.
(A) Cxcl9/Spp1 ratio in MC38 bulk RNA-Seq samples from responder and non-responder strain tumors. **P < 0.01; *P < 0.05; n.s. – not significant (B) Ridge plot depicting the distribution of correlation coefficients between each human homologue of the murine genes with the CXCL9/SPP1 ratio in four TCGA cohort. Gene sets are divided based upon whether each gene is located within or outside our four murine QTL and whether the gene was a DEG or not in bulk RNA-Seq of responder and non-responder strains. The only gene set comprising of genes that are within the QTL and a DEG show a subset highly correlated with CXCL9/SPP1 ratio in human tumors. (C) Cox proportional hazards analysis showing the predictive significance of transcriptional biomarkers derived using the various genesets shown. (D) The predicted tumor immune subtype of N = 23 MC38 tumors is subtype 4. (E) Superior survival is seen in patients undergoing aPD1 treatment who are high expressors (defined as having a score above the median) of our 12 genes-per-QTL transcriptional biomarker (F) Cox proportional hazards analysis showing the predictive significance of the 12 gene-per-QTL transcriptional biomarkers over all other biomarkers when tested on only patients with immune subtype 4 tumors treated with aPD1 therapy. (G) As in (F), but transcriptional biomarkers were applied to patients with immune subtypes 1, 2, 3, or 6 as shown in facets.