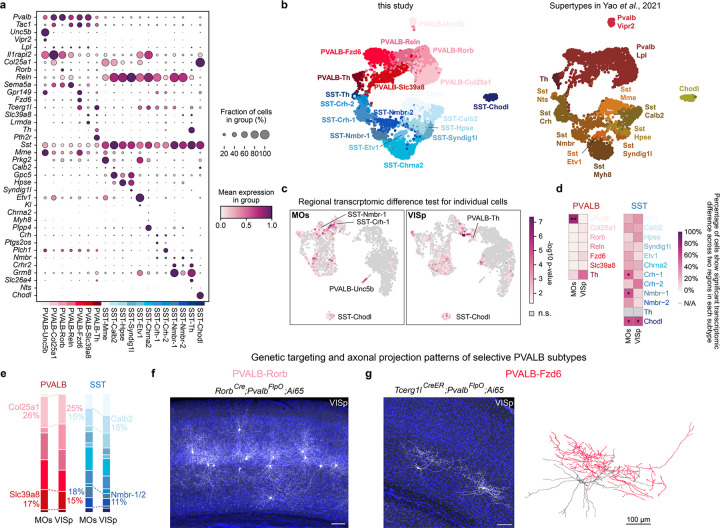
Extended Data Fig. 1. Features of PVALB and SST interneuron subtypes and regional-specific transcriptomic differences.
a, Dot plot showing the expression of marker genes for each PVALB and SST interneuron subtype. b, Analysis of published data showing the correspondence between subtypes identified in this study and supertypes defined in a previous publication2. c, Transcriptomic differences of PVALB and SST interneuron between MOs and VISp regions were assessed by Emergene (See Methods), which calculates a p-value reflecting the enrichment of region-specific gene signatures for individual nuclei, via a permutation test. n.s., not significant. d, Heatmap showing the percentage of cells within each cluster that exhibited significant regional differences. Cell types with ≥ 50% of cells showing regional significance were annotated with the p-value at the 50th percentile for each cell type as calculated by Emergene (*p<0.05; **p< 0.01). N/A: not analyzed, cluster size <10 cells. Detailed p-values are provided in Supplemental Table 2. e, Same bar plot as in Fig. 1b, now fully colored to indicate all subtypes. f, Intersectional genetic strategy preferentially targeting PVALB-Rorb interneurons, showing their axons concentrated in L4. Note that this strategy also labels some L5 PNs in SSp. g, Intersectional genetic strategy preferentially targeting PVALB-Fzd6 interneurons, showing that these interneurons reside in L5b and extend their axons laterally within L5b. Sparse labeling can be achieved via low dose of tamoxifen administration, allowing for the reconstruction of PVALB-Fzd6 interneuron morphology, with one example shown to the right. Note that this strategy also labels L5 PT neurons in SSp. Scale bars: 100 μm.