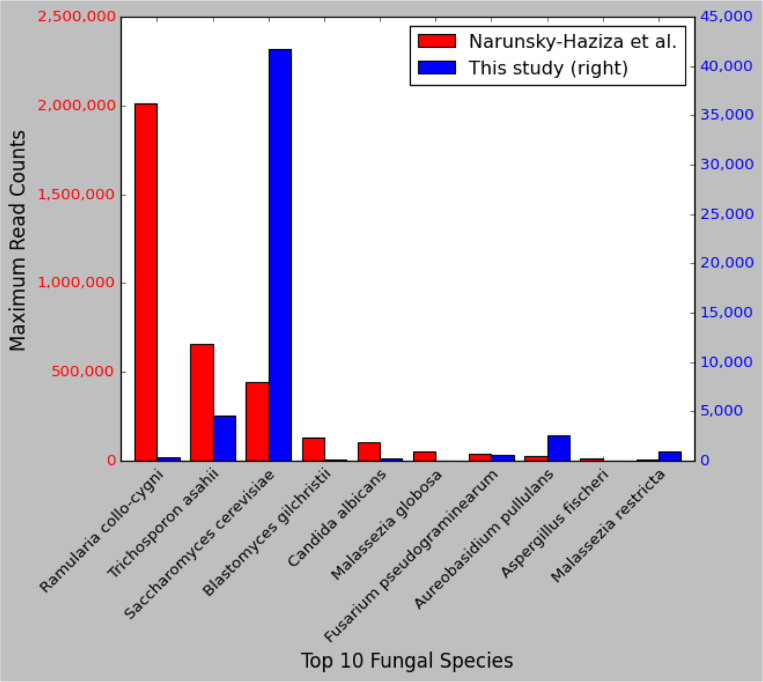
Figure 5.
Comparison of read counts in fungal species. Shown are the top ten most-abundant fungal species ranked by the maximum number of reads reported in Narunsky-Haziza et al. 2, across 4,271 WGS samples from 25 cancer types. The left axis shows read counts as reported by Narunsky-Haziza et al., and the right axis shows read counts as computed in our re-analysis for the same samples and the same species. Note that the y-axis scales differ by a factor of 50. Note also that Narunsky-Haziza et al. reported a maximum of ~441K reads from one sample matching S. cerevisiae, a finding that we were unable to replicate. In our analysis of the same sample using reads filtered to remove human (see Methods), we found ~40K reads matching S. cerevisiae, as shown in the figure. These matches predominantly fell within a region of the yeast genome that contains the 18S rRNA gene and that has significant similarity to human, suggesting that these were human reads that slipped past the filtering step.