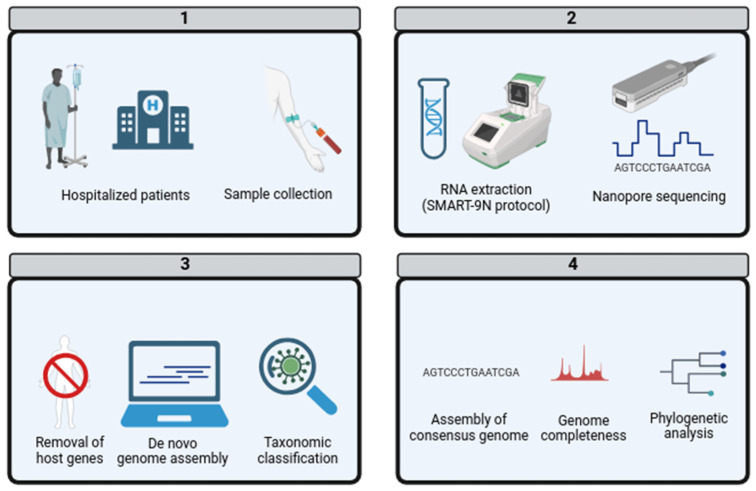
Figure 1.
Metagenomic workflow analysis of AFI and SND in hospitalized patients. (1) Sample collection: Collection of biological samples from hospitalized patients diagnosed with acute febrile illness [AFI] or severe neurological disorder [SND] of unidentified origin. (2) Laboratory Processing: Samples underwent RNA extraction using the SMART-9N protocol, followed by metagenomic sequencing employing nanopore technology. (3) Data Analysis: Sequences were subjected to quality control, the removal of host genetic material, and the generation of contigs through de novo genome assembly. These contigs were then classified taxonomically. (4) Pathogen Identification and Further Analysis: Identified pathogens with coverage exceeding 80% were used to assemble a consensus genome. The completeness of these genomes was assessed, culminating in the construction of a phylogenetic tree.