Figure 1.
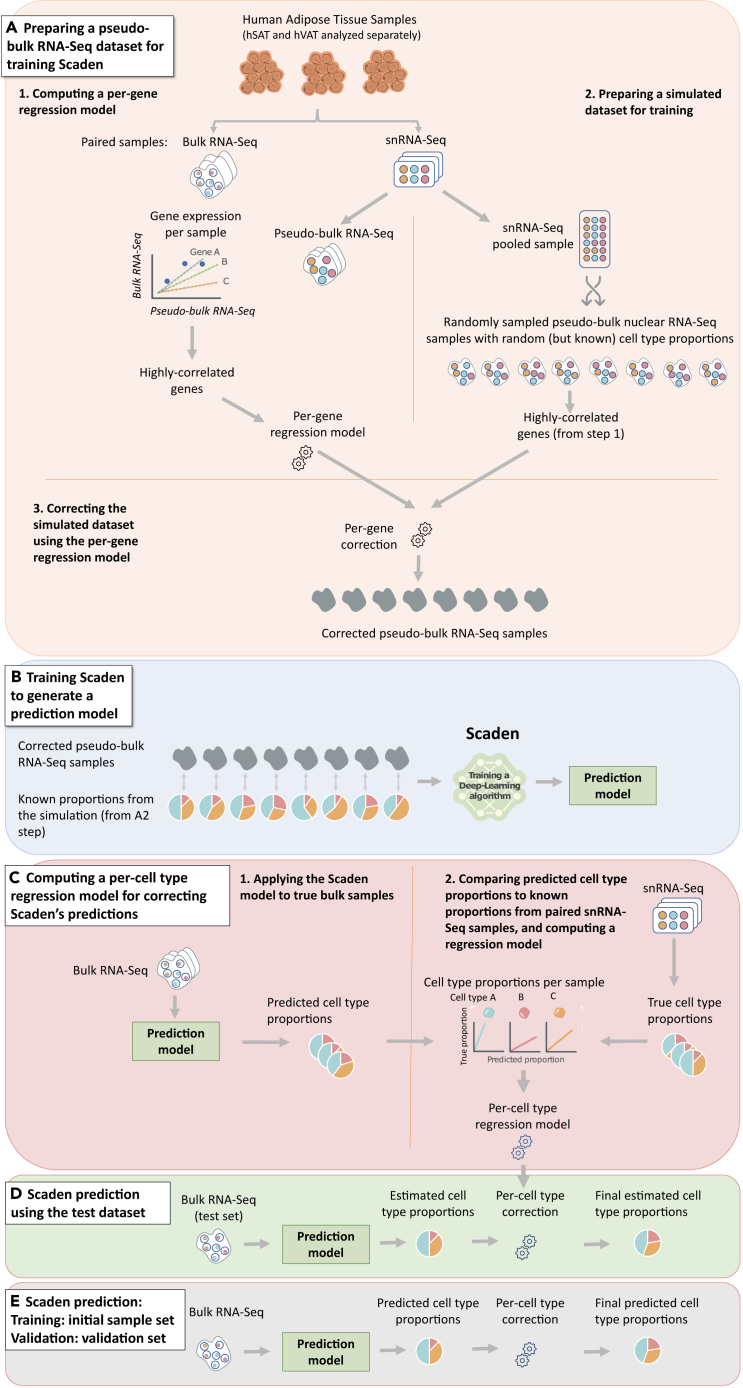
sNucConv workflow
sNucConv is a Scaden-based algorithm developed for estimating cell-type proportions from bulk RNA-seq data while training on snRNA-seq, rather than scRNA-seq, data.
(A–D) denote the 4 stages of sNucConv, which include: (A) the conversion of snRNA-seq dataset into pseudo-bulk training set with per-gene correction, (B) generating a Scaden-based prediction model, (C) per cell-type regression model generation (D) and correction to obtain the final sNucConv deconvolution output.
(E) sNucConv was trained on the initial samples set (7 hVAT and 5 hSAT) and applied onto an independent validation samples set (5 hVAT and 5 hSAT).