Table 4.
Docking simulation results of top two active ingredients against target enzymes EGFR, PI3K, and MAPK(ERK).
| Active Metabolites PubChem Id (CID) |
Target Enzymes | Binding Score kcal/mol |
Key Amino Acid Residues |
Type of Binding | Two-Dimensional Representation |
|---|---|---|---|---|---|
| 442879 | EGFR | −6.2373 | Lys745 CSO797 |
Hydrophobic H-Bonding |
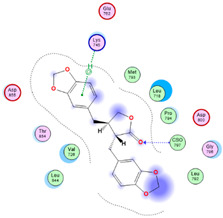
|
| 5312775 | EGFR | −6.1899 | Lys745 Glu762 Asp855 |
H-Bonding H-Bonding H-Bonding |
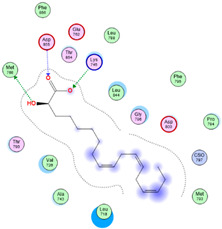
|
| 51683 | PI3K | −5.1383 | Asp810 Asp933 Lys802 |
H-Bonding, Ionic H-Bonding H-Bonding |
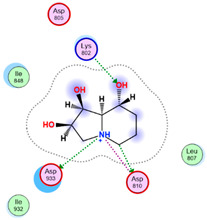
|
| 129846263 | PI3K | −4.8623 | Lys802 Tyr836 |
H-Bonding, Ionic H-Bonding |
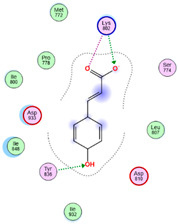
|
| 5312775 | ERK | −4.9854 | Ser170 Asn171 Lys168 Ala52 |
H-Bonding H-Bonding H-Bonding H-Bonding |
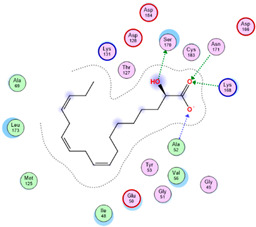
|
| 46178008 | ERK | −4.8738 | Lys168 Asp184 Glu50 |
2 H-Bonding H-Bonding Hydrophobic |
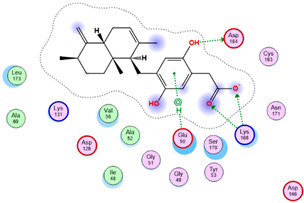
|
