Abstract
Poliovirus infection induces an overall inhibition of host protein synthesis, although some mRNAs continue to be translated, suggesting different translation requirements for cellular mRNAs. It is known that ribosomal protein mRNAs are translationally regulated and that the phosphorylation of ribosomal protein S6 is involved in the regulation. Here, we report that the translation of ribosomal protein mRNAs resists poliovirus infection and correlates with an increase in p70s6k activity and phosphorylation of ribosomal protein S6.
Poliovirus infection results in a drastic shutoff of cellular protein synthesis, accompanied by a selective production of viral proteins (11, 54). This is achieved mostly at the level of translation by specific impairment of the cap-dependent initiation step (17, 28). In fact, the mRNA of poliovirus, and that of other picornaviruses, is uncapped and characterized by a long and structured 5′ untranslated region (5′UTR) where an internal ribosome entry site (IRES) can promote cap-independent translation initiation (19, 22, 43). One of the mechanisms responsible for the inhibition of cap-stimulated translation involves the modification and inactivation of the translation initiation factor eIF4F, due to the cleavage of the eIF4G subunit (11). On the other hand, the eIF4G cleavage products can facilitate the translation initiation of viral RNAs, mediated by an IRES, and of uncapped cellular RNAs (41). Although most of the host protein synthesis is inhibited in poliovirus-infected cells, the translation of some cellular mRNAs occurs. They include the heat shock protein (HSP) mRNAs and the immunoglobulin heavy-chain binding protein, c-myc, and eIF4G mRNAs, which use the mechanisms of internal initiation (25, 39, 50). The cellular modification induced by viral infection to cellular protein synthesis can help to identify the mechanisms that normally control mRNA translation.
We were interested in the regulatory mechanisms that control the translation of ribosomal protein (rp) mRNAs (rp-mRNAs) (1, 37). It is known that the translation of rp-mRNAs is regulated by elements contained in the 5′UTR of rp-mRNAs and, in particular, by a typical terminal oligopyrimidine segment (29, 34). Putative transacting factors can bind the 5′UTR of rp-mRNAs in mammalian and Xenopus cells (5, 26), where they were identified as the La protein and the cellular nucleic acid binding protein (44, 45). Furthermore, it was reported that in mitogen-stimulated cells, the efficiency of translation of mRNAs carrying a 5′-terminal pyrimidine tract is mediated by the activity of p70S6k, the kinase responsible for the phosphorylation of r protein S6 (4, 23, 24, 55).
In this study, we have investigated the behavior of the class of rp-mRNAs under the translational conditions caused by poliovirus infection, in order to obtain information on the mechanisms that control their translation.
Translation of rp-mRNAs during poliovirus infection.
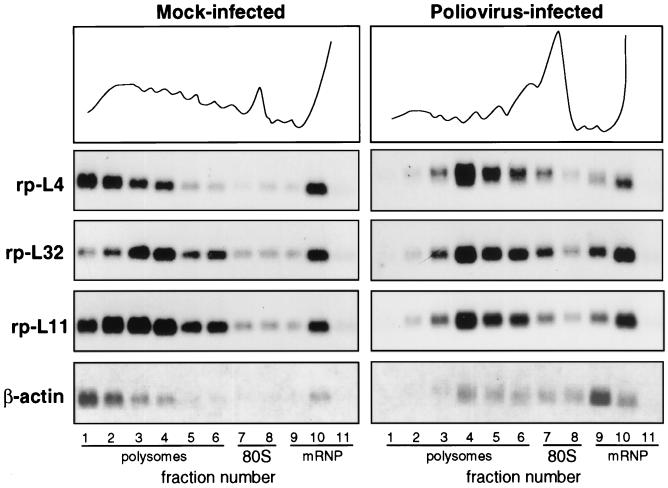
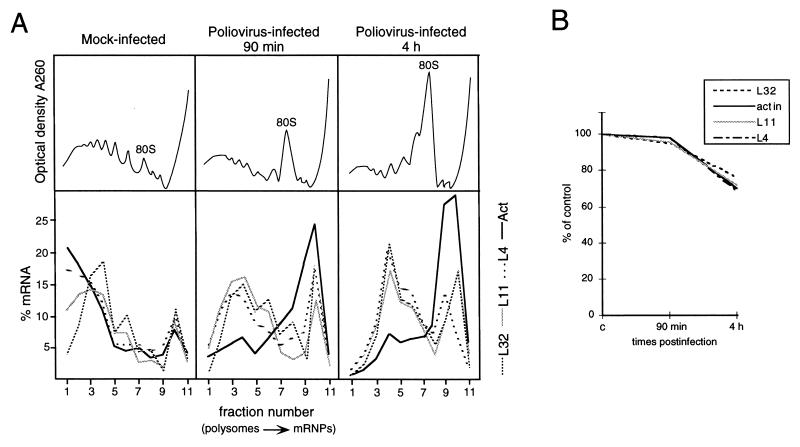
We analyzed the mRNA distribution between polysomes and messenger ribonucleoprotein particles (mRNPs) in mock-infected and poliovirus-infected HEp-2 cells. Cells were infected with the poliovirus type 1 Mahoney strain and incubated for 4 h. Extracts corresponding to one plate of cell culture from mock-infected and poliovirus-infected cells were fractionated by sucrose gradient centrifugation, and the RNA was extracted from the fractions. Amounts corresponding to the same volumes of gradient fractions were analyzed by Northern blotting as previously described (32). A representative example of these experiments is given in Fig. 1, where the polysome-mRNP distribution of rp-mRNAs is compared to that of β-actin mRNA, a control mRNA subjected to shutoff. It should be noticed that in these experiments, only the distribution of the mRNAs along the gradients should be compared between mock-infected and infected cells and not the absolute amount of RNA. In mock-infected cells, about 70 to 80% of the mRNAs analyzed were loaded onto polysomes to be actively translated. In infected cells, β-actin mRNA was mostly displaced to mRNPs at the top of the gradient, as expected, while a large part of the L4, L32, and L11 rp-mRNAs was still associated with polysomes. However, these rp-mRNAs were associated with small polysomes, indicating that in infected cells, translation initiation might be less efficient than in uninfected cells. To obtain further information about the translational behavior of rp-mRNAs at different infection times, we analyzed the polysome-mRNP distribution of L4, L32, L11, and β-actin mRNAs at 90 min and 4 h after infection. Figure 2A shows graphically that, compared with mock-infected cells, the distributions of the rp-mRNAs and β-actin were soon quite different. At 90 min after infection, polysome-associated β-actin mRNA started decreasing and the dislocation of this mRNA to the top of the gradient reached 90% within 4 h. On the contrary, 90 min after infection, about 60% of the L4, L32, and L11 mRNAs was still associated with large polysomes and after 4 h they remained associated with polysomes which, however, were smaller. Since gradient analysis is intended to show the translational activity of the mRNAs and not to quantify their absolute amount, quantitative aliquots of each extract at different times of infection were taken before gradient loading for total RNA analysis by Northern blot hybridization to different probes. The hybridization signals were quantified by comparison with 5S rRNA, which is structured in the ribosomes and therefore is expected to be fairly stable. Figure 2B shows that the level of all the mRNAs analyzed does not change appreciably up to 90 min and, with minor differences, decreases by about 30 to 35% at 4 h, indicating that rp-mRNAs and β-actin mRNA are subjected to similar rates of degradation.
FIG. 1.
Polysome-mRNP distribution of mRNA in mock-infected and poliovirus-infected cells. HEp-2 cells were grown at 37°C in Eagle’s minimum essential medium supplemented with 10% fetal calf serum. When cells reached 80% confluence, the Mahoney strain of poliovirus type 1 was added at a multiplicity of infection of 50 PFU per cell. A 9-cm-diameter plate culture of mock-infected and infected cells was lysed at 4 h postinfection (10 mM NaCl, 10 mM MgCl2, 1% Triton X-100, 1% Na-deoxycholate, 1 mM dithiothreitol, 10 mM Tris-HCl, pH 7.4) to prepare cytoplasmic extracts (32). Cycloheximide, often used to prevent polysome runoff, was not added prior to extract preparation, as this drug sometime creates problems (38). Polysome protection can also be obtained by quickly preparing the extract and loading the sample on the gradient under strict temperature control. The extracts were separated on 15 to 50% sucrose gradients in gradient buffer (0.1 M NaCl, 10 mM MgCl2, 30 mM Tris-HCl, pH 7). Gradient fractions were collected while the optical density profile at 260 nm was monitored (top), and the RNA was prepared by protease K-SDS-phenol extraction (32) of the fractions. The RNAs from equal volumes of mock-infected and infected gradient fractions were loaded onto two gels and analyzed by Northern blotting and autoradiography as previously described (32). Each filter was subsequently hybridized to probes for rp-L4 (2), rp-L32 (10), rp-L11 (12), and β-actin (6) mRNAs to obtain a reliable comparison of the distribution of the various RNAs along the same gradient. Probes were prepared by the random primer technique.
FIG. 2.
Time course of polysome-mRNP distribution and accumulation of mRNAs during poliovirus infection. (A) Mock-infected and poliovirus-infected cells were processed at the times indicated as described in the legend to Fig. 1. Northern blots were subsequently hybridized with rp-L4, rp-L11, rp-L32, and β-actin probes. Measurement of radioactivity, reported as the percentage of mRNA in each fraction, was done by PhosphorImager (Molecular Dynamics) analysis. The optical density profiles of the sucrose gradients were monitored at 260 nm (top), and the positions of the 80S monomers are indicated. These experiments were performed at least three times, and the results were consistently similar. (B) One tenth of each extract, prepared at the indicated time postinfection, was taken before gradient loading. The RNA was extracted, and equivalent amounts were analyzed by Northern blotting as described in the legend to Fig. 1. The filters were subsequently hybridized to rp-L4, rp-L32, rp-L11, β-actin, and 5S RNA probes (46). Measurement of radioactivity was done by PhosphorImager (Molecular Dynamics) analysis, and the values obtained were normalized to the signal of the 5S rRNA probe. The level of each mRNA is expressed as a percentage of the amount measured in mock-infected cells (c).
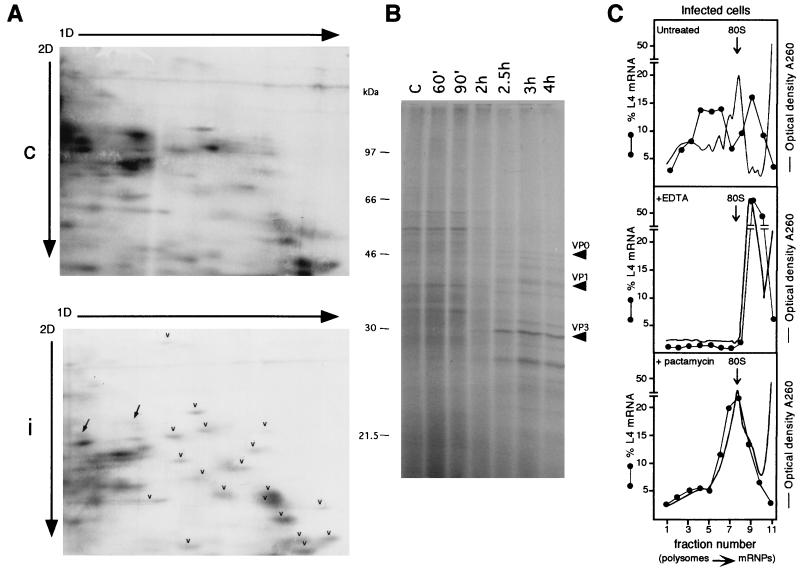
To check the activity of polysome-associated mRNA after poliovirus infection, at 90 min and 3.5 h postinfection, [35S]methionine-[35S]cysteine was added for 30 min to HEp-2 cells to label synthesized r proteins. Thirty minutes of radioactive precursor administration was reasonable for detection of accumulated labeled proteins and to overcome the expected effect of r protein instability due to decreased rRNA synthesis, as will be discussed later. Considering the short labeling time and the fact that more extract could not be loaded onto the gel without pattern distortion, only faint spots can be expected on the two-dimensional (2D) gel, in accordance with data previously obtained with other systems (47). After incubation, protein extracts were prepared from mock-infected and infected cells and analyzed on a 2D gel optimized to resolve ribosomal proteins (58). Gels were Coomassie stained and fluorographed. Radioactive r proteins were identified on the 2D gel by their comigration with the purified human r proteins included in the sample (58). A polysome gradient experiment was performed with an aliquot of the extracts from mock-infected and infected labeled cells to check that the RNA distribution was as expected, namely, as in Fig. 1 (data not shown). At the same time, a protein labeling experiment was performed to check the pattern of total protein synthesis during infection. Figure 3A shows that at 2 h postinfection, the synthesis of r proteins is still efficient (arrowheads), in agreement with the rp-mRNA engagement with polysomes described above. However, at this time, the general inhibition of host cell protein synthesis is already detectable, as shown by the pattern of total protein synthesis at different times after infection (Fig. 3B). Note also the remarkable shutdown of two non-r proteins that can be seen in Fig. 3A (arrows). At 4 h postinfection, r protein labeling is no longer detectable (data not shown). This might be due to the severe inhibition of rRNA synthesis during poliovirus infection (7, 49) that becomes relevant with time, thus preventing accumulation of newly synthesized r proteins. Indeed, it has been observed in other systems that newly synthesized r proteins become unstable when rRNA needed for assembly is not available (8, 47, 58). It was recently reported that, following herpes simplex virus type 1 infection, r proteins continue to be synthesized during protein synthesis shutoff. However, in this system, where 60% rRNA synthesis persists in infected cells, r proteins are stable, as they can assemble with rRNA into ribosomes (52). To ascertain rp-mRNA association with polysomes later in infection as well, some control experiments were carried out. Cytoplasmic extracts were prepared 4 h postinfection and treated with EDTA to dissociate polysomes. Compared to untreated infected cells, a typical rp-mRNA such as L4 was shifted by EDTA treatment to the top of the gradient, implying association with polysomes (Fig. 3C, top and middle). Similar results were obtained for the other rp-mRNAs (data not shown). Moreover, to show that polysome association was due to active translation, cells were treated with pactamycin for 30 min at 3.5 h postinfection. This drug, a translation inhibitor at the initiation step, caused a decrease in polysome size which resulted in the accumulation of L4 mRNA in the dimer and 80S fractions (Fig. 3C, bottom). The same occurred to other rp-mRNAs (data not shown). Rehybridization of the filters with the viral probe showed a signal of the expected size peaking on fraction 3. This might represent the viral particles, which migrate close to, but never coincide with, the rp-mRNAs (data not shown). These results support the hypothesis that rp-mRNAs associate with polysomes, a view further strengthened by the fact that at 2 h postinfection, a similar RNA distribution along the gradient corresponds to protein synthesis activity.
FIG. 3.
Analysis of the activity of polysome-associated RNA in infected cells by metabolic labeling of proteins and by EDTA and pactamycin treatment. (A) HEp-2 cells were grown and infected as described in the legend to Fig. 1. At 90 min postinfection, mock-infected and infected cells were incubated for 30 min with [35S]methionine-[35S]cysteine (Pro-mix; Amersham; >1,000 Ci/mmol) at a concentration of 0.1 mCi/ml. Cells were harvested in phosphate-buffered saline and homogenized in 0.5 ml of ice-cold rp buffer (0.1 M NaCl, 1 mM MgCl2, 10 mM HEPES, pH 7.5), acid extracted (58), and processed by the 2D gel electrophoresis method optimized to resolve r proteins, with the exception that the second-dimension (2D) gel was 13% polyacrylamide (58). A 200-μg sample of r proteins purified from HEp-2 ribosomes (58) was added as markers to an equal amount of control (c) or infected-cell (i) protein extract (200 μg). Gels were fluorographed and exposed to X-ray film for the same time. Arrowheads point to some r proteins, and arrows point to non-r proteins. 1D, first dimension. (B) HEp-2 cells, grown and infected as indicated above, were labeled with Pro-mix (40 μCi/ml) for 10 min at the indicated times after infection. Cells were lysed as described above, and 5 μg of each extract was loaded onto an SDS–12.5% PAGE gel and autoradiographed. Arrows point to viral proteins. Lane C, control. (C) Untreated infected cells (top) were lysed at 4 h postinfection and analyzed on a sucrose gradient as described in the legend to Fig. 1. For the EDTA treatment (middle), cytoplasmic extracts, brought to a concentration of 50 mM EDTA, pH 7.4, were incubated in ice for 5 min, loaded onto sucrose gradients containing 10 mM EDTA instead of magnesium, and analyzed as described in the legend to Fig. 1. To test the effect of pactamycin (bottom), cells were incubated at 3.5 h after infection with 30 ng of pactamycin per ml for 30 min, thus reaching the 4-h infection time of untreated cells, and then processed as described in the legend to Fig. 1. When the gradient fractions were collected, the optical density at 260 nm was monitored. The profiles are shown as a continuous line, and the 80S monomers are indicated by the arrows. Northern blots were hybridized with an rp-L4 probe. Measurement of radioactivity, reported as a percentage of the mRNA in each fraction, was done by PhosphorImager (Molecular Dynamics) analysis.
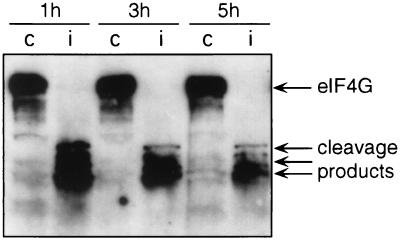
From the results described above, it appears that rp-mRNAs are fairly resistant to protein synthesis shutoff compared to β-actin mRNA. Therefore, in spite of the complete cleavage of eIF4G which already occurs 1 h after infection (Western blot in Fig. 4), rp-mRNAs can still initiate protein synthesis. Similarly, HSP mRNAs are resistant to poliovirus-induced shutoff and it has been proposed that they may utilize cap-independent initiation (48). The limited secondary structure of the HSP 5′UTR, as well as the short and unstructured 5′UTR of rp-mRNAs, may determine a lower dependence on initiation factors compared to more-structured mRNAs (9, 20, 53). In line with this, it has been reported that the efficiency of translation of rp-mRNAs is regulated independently of the level or activity of eIF4E (51), whereas the selective translational repression of mRNAs bearing extensive secondary structure in the 5′UTR is relieved by the overexpression of this factor (27). It was recently reported that eIF4GII, a functional homolog of eIF4G (hence called eIF4GI), can persist longer in poliovirus-infected cells, as shown by the fact that about 30% of the entire form still persists at up to 2 h after infection (14, 15). It can be argued that the rp-mRNA association with polysomes and the r protein synthesis described here could be sustained by eIF4GII in infected cells. However, later in infection, when eIF4GII is completely cleaved, rp-mRNAs are still associated with polysomes, suggesting that other elements are also involved.
FIG. 4.
eIF4G cleavage in poliovirus-infected cells. Cytoplasmic extracts from mock-infected (lanes c) and poliovirus-infected (lanes i) cells were prepared at the times indicated as described in the legend to Fig. 1. Aliquots of the cytoplasmic extracts were precipitated with acetone for protein analysis by SDS-PAGE and Western blotting using the anti-eIF4G antibody as previously described (18).
Analysis of p70S6k activity and S6 phosphorylation during poliovirus infection.
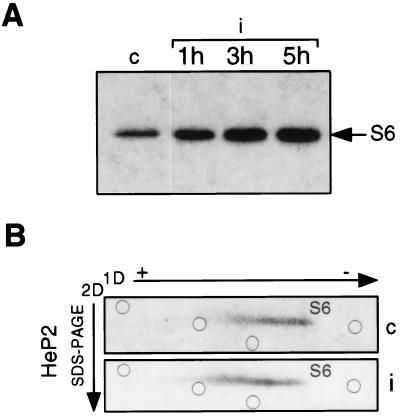
As mentioned above, a relationship among translation of rp-mRNAs, activity of p70S6k, and phosphorylation of r-protein S6 has been reported (23, 24, 55). To investigate the state of p70S6k activity after poliovirus infection, we performed in vitro kinase assays by using p70S6k immunoprecipitated from equal amounts of mock-infected and infected-cell extracts and 40S ribosomal subunits as a substrate (16). As exemplified by the experiment of Fig. 5A, the ability of p70S6k to phosphorylate S6 was maintained and even increased 1 h after infection, reaching a level of two- to three-fold at 5 h. Similar results were consistently obtained by either immunoprecipitation or direct incubation with 40S ribosomes of equal amounts of control and infected-cell extracts. Although we cannot determine whether this was due to higher p70S6k activity or to an increased amount of it, the experiments reproducibly showed an increase in the capacity of p70S6k to phosphorylate S6 in infected cells. Then, to analyze S6 phosphorylation in vivo, cells were labeled with [32P]orthophosphate for 90 min at 2.5 h postinfection, thus reaching a total infection time of 4 h, when the in vitro phosphorylating activity was still increasing (Fig. 5A). Proteins were analyzed by 2D gel electrophoresis as previously described (33), with the exception that in the second dimension, a sodium dodecyl sulfate (SDS)–15% polyacrylamide gel electrophoresis (PAGE) gel was used. The 2D gel electrophoresis conditions were set up to map S6 phosphorylated forms, as indicated in the legend to Fig. 5B. In this system, the hyperphosphorylated forms migrate slower in the first dimension. Figure 5B shows two identical gels loaded with extracts from [32P]orthophosphate-labeled control and infected cells. Compared to control cells, infected cells show a slight shift of the S6 spot toward the anode, as measured by the relative positions of the radioactive S6 and stained, purified r proteins included in each sample. A small decrease in hypophosphorylated forms and a small increase in hyperphosphorylated forms can be seen. This finding, observed in repeated experiments, suggests that in infected cells, the level of S6 phosphorylation is maintained and indeed slightly increased, compared to that in control cells, in line with the result of the p70S6k kinase assay. Similarly, the activity of the S6 kinase and S6 phosphorylation are stimulated following herpes simplex virus type 1 infection (21, 35).
FIG. 5.
p70S6k activity and S6 phosphorylation during poliovirus infection. (A) S100 cytoplasmic extracts from mock-infected (lane c) and poliovirus-infected (i) cells were prepared at the times indicated in the presence of a phosphatase inhibitor as previously described (40). Protein concentration was determined by the Bio-Rad Protein Assay kit. Eight-microgram extract samples were used for immunoprecipitation of p70S6k with M6 antibody, and the immunocomplex was assayed for in vitro kinase activity in the presence of 40S ribosomes at an A260 of 0.45 U as previously described (16). Proteins were separated by SDS–PAGE and autoradiographed. Phosphorylated S6 protein is indicated by the arrow. (B) Mock-infected and poliovirus-infected cells (c and i, respectively) were labeled at 2.5 h after infection in a phosphate-free medium with 20 μCi of [32P]orthophosphate per ml for 90 min. Ribosomes from labeled cells were purified, and ribosomal proteins were extracted in the presence of 40 mM β-glycerophosphate (58). Proteins were analyzed by 2D electrophoresis as previously described (33), with the exception that the second dimension (2D) was an SDS–15% PAGE gel. A 200-μg sample of unlabeled purified HEp-2 ribosomal proteins was added to each sample before gel loading for Coomassie staining and used as position markers in the gel. The positions of stained ribosomal proteins in the S6 area are indicated by open circles. To standardize 2D gel electrophoresis conditions and to map the positions of S6 phosphorylated forms, preliminary experiments were carried out by loading gels with purified 40S subunits phosphorylated in in vitro kinase assays by extracts from quiescent and serum-stimulated Swiss 3T3 cells (16). The level of S6 phosphorylation by quiescent cell extracts was very low and increased 20-fold after serum stimulation. Accordingly, an evident shift in migration of S6 in the 2D gel was observed (data not shown). 1D, first dimension.
It is known that both p70S6k activation and the phosphorylation of the initiation factor 4E binding protein (4E-BP1) are mediated by the mTOR-FRAP signalling pathway (3, 31, 57). The different phosphorylation state of 4E-BP1 affects eIF4E activity, since the dephosphorylated form can sequester eIF4E, thus blocking cap-dependent initiation (30, 42). It has been recently proposed that the p70S6k–4E-BP1 phosphorylation pathway bifurcates immediately upstream from p70S6k (56). Interestingly, it has been found that 4E-BP1 becomes dephosphorylated after poliovirus infection (13) while our data indicate that p70S6k activity and S6 phosphorylation do not decrease but are maintained and somewhat stimulated in infected cells. If this is the case, these observations might suggest that poliovirus infection either influences the p70S6k–4E-BP1 kinase pathway downstream from the bifurcation or differentially regulates specific phosphatases.
In conclusion, we have identified a new class of cellular mRNAs that, besides HSP (39), and some IRES-containing cellular mRNAs recently analyzed with an approach similar to the one used in this study (25), can be translated in poliovirus-infected cells. Moreover, we have shown that the selective translation resistance of rp-mRNAs correlates with the maintenance of p70S6k activity and with a small, but consistent, phosphorylation increase in r protein S6. It is hard to believe that r protein synthesis resistance is an advantage for the virus, as rRNA synthesis is inhibited and ribosome assembly is not possible. It is more likely that r protein mRNA translation can continue because the conditions required are still sufficient, in spite of the drastic damage to cellular translation initiation generated by the infection. This translation survival, that so far can be identified as lower dependence on initiation factors, may reveal a feature of the normal mechanism governing rp-mRNA translation in the cell. It is possible to speculate that this mechanism can be due to the short and typical rp-mRNA 5′UTR; to the utilization of specific ribosomes containing hyperphosphorylated S6, a feature that appears to persist under infection conditions; and to auxiliary factors that specifically bind the 5′UTR of rp-mRNAs (44, 45). Interestingly, one of these binding factors, La protein, is known to have a positive role in poliovirus RNA translation in vitro (36). Study of the mechanisms that govern cellular mRNA resistance to poliovirus-induced shutoff of protein synthesis adds to our knowledge of the cellular response to viral infection and should provide important clues to understand the translational regulation of mRNAs.
Acknowledgments
We thank L. Carrasco for the anti-eIF4G antibody, G. Thomas for the M6 anti-p70S6k antibody, and Pharmacia & Upjohn and M. Kleijn for pactamycin. We also thank F. Amaldi, G. Giannini, and F. Loreni for their critical reading of the manuscript.
This work was partially supported by the EC-DGXII Biotech Program and by Progetto Strategico CNR “Posttranscriptional Controls of Gene Expression.”
REFERENCES
- 1.Amaldi F, Pierandrei-Amaldi P. TOP genes: a translationally controlled class of genes including those coding for ribosomal proteins. In: Jeanteur P, editor. Cytoplasmic fate of eukaryotic mRNA. Vol. 18. Heidelberg, Germany: Springer-Verlag; 1997. pp. 1–17. [DOI] [PubMed] [Google Scholar]
- 2.Bagni C, Mariottini P, Annesi F, Amaldi F. Human ribosomal protein L4: cloning and sequencing of the cDNA and primary structure of the protein. Biochim Biophys Acta. 1993;1216:475–478. doi: 10.1016/0167-4781(93)90017-8. [DOI] [PubMed] [Google Scholar]
- 3.Beretta L, Gingras A C, Svitkin Y V, Hall M N, Sonenberg N. Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. EMBO J. 1996;15:658–664. [PMC free article] [PubMed] [Google Scholar]
- 4.Brown E J, Schreiber S L. A signaling pathway to translational control. Cell. 1996;86:517–520. doi: 10.1016/s0092-8674(00)80125-7. [DOI] [PubMed] [Google Scholar]
- 5.Cardinali B, Di Cristina M, Pierandrei-Amaldi P. Interaction of proteins with the mRNA for ribosomal protein L1 in Xenopus: structural characterization of in vivo complexes and identification of proteins that bind in vitro to its 5′UTR. Nucleic Acids Res. 1993;21:2301–2308. doi: 10.1093/nar/21.10.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cleveland D W, Lopata M A, MacDonald R J, Cowan N J, Rutter W J, Kirschner M W. Number and evolutionary conservation of alpha- and beta-tubulin and cytoplasmic beta- and gamma-actin genes using specific cloned cDNA probes. Cell. 1980;20:95–105. doi: 10.1016/0092-8674(80)90238-x. [DOI] [PubMed] [Google Scholar]
- 7.Contreras G, Summers D F, Maizel J V, Ehrenfeld E. HeLa cell nucleolar RNA synthesis after poliovirus infection. Virology. 1973;53:120–129. doi: 10.1016/0042-6822(73)90471-6. [DOI] [PubMed] [Google Scholar]
- 8.Craig N, Perry R P. Persistent cytoplasmic synthesis of ribosomal proteins during the selective inhibition of ribosome RNA synthesis. Nat New Biol. 1971;229:75–80. doi: 10.1038/newbio229075a0. [DOI] [PubMed] [Google Scholar]
- 9.Degener A M, Silveira Carneiro J, Cassetti C, Pierangeli A, Pagnotti P, Bucci M, Perez Bercoff R. Role of the pyrimidine-rich tract on the translation of ‘chimeric’ polio-hepatitis A mRNAs with engineered 5′-terminal untranslated regions. Virus Res. 1995;37:291–303. doi: 10.1016/0168-1702(95)00030-t. [DOI] [PubMed] [Google Scholar]
- 10.Dudov K P, Perry R P. The gene family encoding the mouse ribosomal protein L32 contains a uniquely expressed intron-containing gene and an unmutated processed gene. Cell. 1984;37:457–468. doi: 10.1016/0092-8674(84)90376-3. [DOI] [PubMed] [Google Scholar]
- 11.Ehrenfeld E. Initiation of translation by picornavirus RNAs. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 549–573. [Google Scholar]
- 12.Filipenko, M., and F. Amaldi. Unpublished data.
- 13.Gingras A C, Svitkin Y, Belsham G J, Pause A, Sonenberg N. Activation of the translational suppressor 4E-BP1 following infection with encephalomyocarditis virus and poliovirus. Proc Natl Acad Sci USA. 1996;93:5578–5583. doi: 10.1073/pnas.93.11.5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gradi A, Imataka H, Svitkin Y V, Rom E, Raught B, Morino S, Sonenberg N. A novel functional human eukaryotic translation initiation factor 4G. Mol Cell Biol. 1998;18:334–342. doi: 10.1128/mcb.18.1.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gradi A, Svitkin Y V, Imataka H, Sonenberg N. Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shut off of host protein synthesis after poliovirus infection. Proc Natl Acad Sci USA. 1998;95:11089–11094. doi: 10.1073/pnas.95.19.11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han J W, Pearson R B, Dennis P B, Thomas G. Rapamycin, wortmannin, and the methylxanthine SQ20006 inactivate p70s6k by inducing dephosphorylation of the same subset of sites. J Biol Chem. 1995;270:21396–21403. doi: 10.1074/jbc.270.36.21396. [DOI] [PubMed] [Google Scholar]
- 17.Helentjaris T, Ehrenfeld E. Control of protein synthesis in extracts from poliovirus-infected cells. I. mRNA discrimination by crude initiation factors. J Virol. 1978;26:510–521. doi: 10.1128/jvi.26.2.510-521.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Irurzun A, Sanchez-Palomino S, Novoa I, Carrasco L. Monensin and nigericin prevent the inhibition of host translation by poliovirus, without affecting p220 cleavage. J Virol. 1995;69:7453–7460. doi: 10.1128/jvi.69.12.7453-7460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jackson R J, Howell M T, Kaminski A. The novel mechanism of initiation of picornavirus RNA translation. Trends Biochem Sci. 1990;15:477–483. doi: 10.1016/0968-0004(90)90302-r. [DOI] [PubMed] [Google Scholar]
- 20.Jackson R J, Kaminski A. Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA. 1995;1:985–1000. [PMC free article] [PubMed] [Google Scholar]
- 21.Jakubowicz T, Leader D P. Activation of a ribosomal protein S6 kinase in mouse fibroblasts during infection with herpesvirus. Eur J Biochem. 1987;168:371–376. doi: 10.1111/j.1432-1033.1987.tb13429.x. [DOI] [PubMed] [Google Scholar]
- 22.Jang S K, Kräusslich H-G, Nicklin M J H, Duke G M, Palmenberg A C, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jefferies H B, Fumagalli S, Dennis P B, Reinhard C, Pearson R B, Thomas G. Rapamycin suppresses 5′TOP mRNA translation through inhibition of p70s6k. EMBO J. 1997;16:3693–3704. doi: 10.1093/emboj/16.12.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jefferies H B, Reinhard C, Kozma S C, Thomas G. Rapamycin selectively represses translation of the ‘polypyrimidine tract’ mRNA family. Proc Natl Acad Sci USA. 1994;91:4441–4445. doi: 10.1073/pnas.91.10.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johannes G, Sarnow P. Cap-independent polysomal association of natural mRNAs encoding c-myc, BIP, and eIF4G conferred by internal ribosome entry sites. RNA. 1998;4:1500–1513. doi: 10.1017/s1355838298981080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaspar R L, Kakegava T, Cranston H, Morris D R, White M W. A regulatory cis element and a specific binding factor involved in the mitogenic control of murine ribosomal protein L32 translation. J Biol Chem. 1992;267:508–514. [PubMed] [Google Scholar]
- 27.Koromilas A E, Lazaris-Karatzas A, Sonenberg N. mRNAs containing extensive secondary structure in their 5′ non-coding region translate efficiently in cells overexpressing initiation factor eIF-4E. EMBO J. 1992;11:4153–4158. doi: 10.1002/j.1460-2075.1992.tb05508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leibowitz R, Penman S. Regulation of protein synthesis in HeLa cells. III. Inhibition during poliovirus infection. J Virol. 1971;8:661–668. doi: 10.1128/jvi.8.5.661-668.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levy S, Avni D, Hariharan N, Perry R P, Meyuhas O. Oligopyrimidine tract at the 5′ end of mammalian ribosomal protein mRNAs is required for their translational control. Proc Natl Acad Sci USA. 1991;88:3319–3323. doi: 10.1073/pnas.88.8.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin T A, Kong X, Haystead T A, Pause A, Belsham G, Sonenberg N, Lawrence J C., Jr PHAS-I as a link between mitogen-activated protein kinase and translation initiation. Science. 1994;266:653–656. doi: 10.1126/science.7939721. [DOI] [PubMed] [Google Scholar]
- 31.Lin T A, Kong X, Saltiel A R, Blackshear P J, Lawrence J C., Jr Control of PHAS-I by insulin in 3T3-L1 adipocytes. Synthesis, degradation, and phosphorylation by a rapamycin-sensitive and mitogen-activated protein kinase-independent pathway. J Biol Chem. 1995;270:18531–18538. doi: 10.1074/jbc.270.31.18531. [DOI] [PubMed] [Google Scholar]
- 32.Loreni F, Amaldi F. Translational regulation of ribosomal protein synthesis in Xenopus cultured cells: mRNA relocation between polysomes and RNP during nutritional shifts. Eur J Biochem. 1992;205:1027–1032. doi: 10.1111/j.1432-1033.1992.tb16870.x. [DOI] [PubMed] [Google Scholar]
- 33.Madjar J J, Michel S, Cozzone A J, Reboud J P. A method to identify individual proteins in four different two-dimensional gel electrophoresis systems: application to Escherichia coli ribosomal proteins. Anal Biochem. 1979;92:174–182. doi: 10.1016/0003-2697(79)90641-9. [DOI] [PubMed] [Google Scholar]
- 34.Mariottini P, Amaldi F. The 5′ untranslated region of mRNA for ribosomal protein S19 is involved in its translational regulation during Xenopus development. Mol Cell Biol. 1990;10:816–822. doi: 10.1128/mcb.10.2.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masse T, Garcin D, Jacquemont B, Madjar J J. Herpes simplex virus type-1-induced stimulation of ribosomal protein S6 phosphorylation is inhibited in neomycin-treated human epidermoid carcinoma 2 cells and in ras-transformed cells. Eur J Biochem. 1990;194:287–291. doi: 10.1111/j.1432-1033.1990.tb19455.x. [DOI] [PubMed] [Google Scholar]
- 36.Meerovitch K, Svitkin Y V, Lee H S, Lejbkowicz F, Kenan D J, Chan E K L, Agol V I, Keene J D, Sonenberg N. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J Virol. 1993;67:3798–3807. doi: 10.1128/jvi.67.7.3798-3807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meyuhas O, Avni D, Shama S. Translational control of ribosomal protein mRNAs in eukaryotes. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 363–388. [Google Scholar]
- 38.Meyuhas O, Bibrerman Y, Pierandrei-Amaldi P, Amaldi F. Analysis of polysomal RNA. In: Krieg P A, editor. A laboratory guide to RNA. Isolation, analysis and synthesis. New York, N.Y: Wiley-Liss; 1996. pp. 65–81. [Google Scholar]
- 39.Munoz A, Alonso M A, Carrasco L. Synthesis of heat-shock proteins in HeLa cells: inhibition by virus infection. Virology. 1984;137:150–159. doi: 10.1016/0042-6822(84)90018-7. [DOI] [PubMed] [Google Scholar]
- 40.Novak-Hofer I, Thomas G. An activated S6 kinase in extracts from serum- and epidermal growth factor-stimulated Swiss 3T3 cells. J Biol Chem. 1984;259:5995–6000. [PubMed] [Google Scholar]
- 41.Ohlmann T, Rau M, Pain V M, Morley S J. The C-terminal domain of eukaryotic protein synthesis initiation factor (eIF) 4G is sufficient to support cap-independent translation in the absence of eIF4E. EMBO J. 1996;15:1371–1382. [PMC free article] [PubMed] [Google Scholar]
- 42.Pause A, Belsham G J, Gingras A C, Donze O, Lin T A, Lawrence J C, Jr, Sonenberg N. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature. 1994;371:762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 43.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 44.Pellizzoni L, Cardinali B, Lin-Marq N, Mercanti D, Pierandrei-Amaldi P. A Xenopus laevis homologue of the La autoantigen binds the pyrimidine tract of the 5′UTR of ribosomal protein mRNAs in vitro: implication of a protein factor in complex formation. J Mol Biol. 1996;259:904–915. doi: 10.1006/jmbi.1996.0368. [DOI] [PubMed] [Google Scholar]
- 45.Pellizzoni L, Lotti F, Maras B, Pierandrei-Amaldi P. Cellular nucleic acid binding protein binds a conserved region of the 5′UTR of Xenopus laevis ribosomal protein mRNAs. J Mol Biol. 1997;267:264–275. doi: 10.1006/jmbi.1996.0888. [DOI] [PubMed] [Google Scholar]
- 46.Pieler T, Appel B, Oei S L, Mentrel H, Erdmann A. Point mutational analysis of the Xenopus laevis 5S gene promoter. EMBO J. 1985;4:1847–1853. doi: 10.1002/j.1460-2075.1985.tb03859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pierandrei-Amaldi P, Beccari E, Bozzoni I, Amaldi F. Ribosomal protein production in normal and anucleolate Xenopus embryos: regulation at the posttranscriptional and translational levels. Cell. 1985;42:317–323. doi: 10.1016/s0092-8674(85)80127-6. [DOI] [PubMed] [Google Scholar]
- 48.Rhoads R E, Lamphear B J. Cap-independent translation of heat shock messenger RNAs. Curr Top Microbiol Immunol. 1995;203:131–153. doi: 10.1007/978-3-642-79663-0_7. [DOI] [PubMed] [Google Scholar]
- 49.Rubinstein S J, Dasgupta A. Inhibition of rRNA synthesis by poliovirus: specific inactivation of transcription factors. J Virol. 1989;63:4689–4696. doi: 10.1128/jvi.63.11.4689-4696.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sarnow P. Translation of glucose-regulated protein 78/immunoglobulin heavy-chain binding protein mRNA is increased in poliovirus-infected cells at a time when cap-dependent translation of cellular mRNAs is inhibited. Proc Natl Acad Sci USA. 1989;86:5795–5799. doi: 10.1073/pnas.86.15.5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shama S, Avni D, Frederickson R M, Sonenberg N, Meyuhas O. Overexpression of initiation factor eIF-4E does not relieve the translational repression of ribosomal protein mRNAs in quiescent cells. Gene Expr. 1995;4:241–252. [PMC free article] [PubMed] [Google Scholar]
- 52.Simonin D, Diaz J J, Masse T, Madjar J J. Persistence of ribosomal protein synthesis after infection of HeLa cells by herpes simplex virus type 1. J Gen Virol. 1997;78:435–443. doi: 10.1099/0022-1317-78-2-435. [DOI] [PubMed] [Google Scholar]
- 53.Sonenberg N. Cap-binding proteins of eukaryotic messenger RNA: functions in initiation and control of translation. Prog Nucleic Acid Res Mol Biol. 1988;35:173–207. doi: 10.1016/s0079-6603(08)60614-5. [DOI] [PubMed] [Google Scholar]
- 54.Sonenberg N, Meerovitch K. Translation of poliovirus mRNA. Enzyme. 1990;44:278–291. doi: 10.1159/000468765. [DOI] [PubMed] [Google Scholar]
- 55.Terada N, Patel H R, Takase K, Kohno K, Nairn A C, Gelfand E W. Rapamycin selectively inhibits translation of mRNAs encoding elongation factors and ribosomal proteins. Proc Natl Acad Sci USA. 1994;91:11477–11481. doi: 10.1073/pnas.91.24.11477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.von Manteuffel S R, Dennis P B, Pullen N, Gingras A-C, Sonenberg N, Thomas G. The insulin-induced signalling pathway leading to S6 and initiation factor 4E binding protein 1 phosphorylation bifurcates at a rapamycin-sensitive point immediately upstream of p70s6k. Mol Cell Biol. 1997;17:5426–5436. doi: 10.1128/mcb.17.9.5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.von Manteuffel S R, Gingras A C, Ming X F, Sonenberg N, Thomas G. 4E-BP1 phosphorylation is mediated by the FRAP-p70s6k pathway and is independent of mitogen-activated protein kinase. Proc Natl Acad Sci USA. 1996;93:4076–4080. doi: 10.1073/pnas.93.9.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Warner J R. In the absence of ribosomal RNA synthesis, the ribosomal proteins of HeLa cells are synthesized normally and degraded rapidly. J Mol Biol. 1977;115:315–333. doi: 10.1016/0022-2836(77)90157-7. [DOI] [PubMed] [Google Scholar]