Abstract
Human prion diseases are rare, transmissible and often rapidly progressive dementias. The most common type, sporadic Creutzfeldt-Jakob disease (sCJD), is highly variable in clinical duration and age at onset. Genetic determinants of late onset or slower progression might suggest new targets for research and therapeutics. We assembled and array genotyped sCJD cases diagnosed in life or at autopsy. Clinical duration (median:4, interquartile range (IQR):2.5–9 (months)) was available in 3,773 and age at onset (median:67, IQR:61–73 (years)) in 3,767 cases. Phenotypes were successfully transformed to approximate normal distributions allowing genome-wide analysis without statistical inflation. 53 SNPs achieved genome-wide significance for the clinical duration phenotype; all of which were located at chromosome 20 (top SNP rs1799990, pvalue = 3.45x10-36, beta = 0.34 for an additive model; rs1799990, pvalue = 9.92x10-67, beta = 0.84 for a heterozygous model). Fine mapping, conditional and expression analysis suggests that the well-known non-synonymous variant at codon 129 is the obvious outstanding genome-wide determinant of clinical duration. Pathway analysis and suggestive loci are described. No genome-wide significant SNP determinants of age at onset were found, but the HS6ST3 gene was significant (pvalue = 1.93 x 10−6) in a gene-based test. We found no evidence of genome-wide genetic correlation between case-control (disease risk factors) and case-only (determinants of phenotypes) studies. Relative to other common genetic variants, PRNP codon 129 is by far the outstanding modifier of CJD survival suggesting only modest or rare variant effects at other genetic loci.
Introduction
Human prion diseases are rare and often rapidly progressive dementia disorders with no known treatments that slow the disease process. The most common type, sporadic Creutzfeldt-Jakob disease (sCJD), occurs at a relatively uniform annual incidence of 1-2/million, equating to a lifetime risk of approximately 1:5000 [1]. The clinical presentation and progression of the disorder is remarkably variable both in terms of the initial symptoms and signs, age at onset and clinical duration [2–4]. Patients typically present in late middle or old age but have been reported in adolescence and early adulthood, and at the extremes of old age [5–7]. The median clinical duration is usually reported as five months with a range of only a few weeks to several years [2]. Ability to estimate the likely clinical duration could help with timely decisions about care [8].
Prions are proteinaceous pathogens formed of host prion protein (PrP) which cause mammalian prion diseases like bovine spongiform encephalopathy, sheep scrapie, chronic wasting disease of cervids, and the human disorders [9]. The recently determined structures of mouse and hamster prions reveals assemblies of PrP in a parallel in-register beta sheet structure with two domains [10, 11], in marked contrast to the predominant alpha-helices of normal cellular PrP [12]. Prions are thought to replicate by a process of binding of normal cellular PrP, conformational change and subsequently aggregate fission. In several model systems, incubation time of prion disease is influenced by PrP gene expression, primary sequence and polymorphisms, as well as prion strains [13], thought to be conferred by structural variation of the pathogen [14]. Experiments using animal or cellular model systems have led to proposals of several possible non-PrP mechanisms of toxicity in prion diseases, involving PrP binding partners on the cell surface and downstream intracellular changes [15–17]; however, their relevance to the human diseases is yet to be determined.
Human epidemiological and genetic studies have identified factors that associate with survival time in sCJD [2, 8, 18, 19], including demographic factors, prion protein genotype, molecular strain typing of protease-resistant prion protein by Western blot analysis, and a range of biofluid, tissue, imaging, and neurophysiological biomarkers [20]. Many biomarkers simply measure the rate or extent of neuronal injury, loss, or dysfunction, or immune cell or glial responses, whereas genetic associations are implicitly causal of modified clinical phenotypes. In this study, we sought to determine the effects of genome-wide common genetic variation on key clinical phenotypes of sCJD, to develop evidence of modifiers relevant to human prion diseases that might benefit understanding of disease processes and generate new ideas for therapeutics.
Materials and methods
Diagnosis and clinical phenotypes
Details of the contributing sites and diagnostic criteria were given in a previous publication [19]. In short, all patient participants were deceased and gained a diagnosis in life of probable CJD or definite CJD after a post-mortem examination (using contemporary epidemiological criteria which changed over the recruitment period 1990–2019). “Probable CJD” is an epidemiological term that now equates to an almost certain diagnosis of CJD post-mortem (e.g. [21]). Age at clinical onset was given to the nearest month. Clinical duration was based on the examining physician’s impression of the date of onset of the first symptom that subsequently was thought to be a component of the disease syndrome until death in months.
Samples used in this study were obtained over several decades and the data were accessed from January 2023 until now.
Genotyping and quality control
In addition to 4110 samples previously reported, genotyped on an Illumina OmniExpress array [19], 819 new samples were genotyped using Illumina’s Global Screening Array. Standard sample and genotyping quality control was performed using PLINK v1.90b3v, which generated 6,308,901 autosomal SNPs of high quality. Samples with a call rate below 98% and population outliers identified via multidimensional scaling were removed. Additionally, related samples (Pi_Hat > 0.1875) were discarded. Only autosomal SNPs with a genotyping rate of >99%, a minor allele frequency ≥ 0.01 and SNPs not deviating from the Hardy-Weinberg equilibrium (P>10−4) were retained. SNPs of A/T or G/C transversion or those which showed deviation from heterozygosity mean (±3 SD) were excluded. To ensure consistency with the Michigan Imputation Server pipeline the target VCF files were checked against the 1000 Genomes Project reference panel (https://faculty.washington.edu/browning/conform-gt.html/). Genotypes were imputed using the Michigan Imputation Server (using Minimac4 assuming a mixed population, HRC r1.1 2016 (Haplotype Reference Consortium) as reference panel and Eagle 2.4 for phasing) [22]. A post-imputation QC analysis was carried out and SNPs with an r2 threshold lower than 0.3 (removing 70% of poorly imputed SNPs) were excluded.
Statistical analysis
SNPTEST (v2.5.2) was used to perform association and conditional analysis with an additive and heterozygous logistic regression model, using sex, contributing site and 10 population covariates generated with PLINK (v1.90b3v; www.cog-genomics.org/plink/1.9/). Genetic correlation between this (using duration as phenotype) and the previously conducted sCJD case-control study [19] was performed using LDSC [23], a software tool for linkage disequilibrium (LD) score and heritability estimation using summary statistics. Meta-analysis was performed using METAL combining the previously published GWAS case-control data [19] and the case-only data described here using summary test statistics as input (6,314,883 SNPs in the union list) and adopting the sample-based approach by combining z-scores across samples in a weighted sum proportional to study sample sizes. FUMA [24], using an integrated Magma gene-based and gene-set analysis on the GWAS summary data, was utilised to perform pathway analysis to identify genes and pathways associated with sCJD risk. FUMA also provides information about chromatin interaction, expression patterns and shared molecular functions between genes. MAGMA software was also utilised for gene-based / gene-set analysis [25]. Power analysis was performed using R functions taken from the Github site https://github.com/kaustubhad/gwas-power provided by Kaustubh Adhikari (UCL Division of Biosciences, University College London).
Ethics
The research project has approval from the NHS Health Research authority (London—Harrow Research Ethics Committee, London, UK); the REC reference is 05/Q0505/113. Written informed consent has been obtained.
Results
We performed the association analysis with 3773 (duration as phenotype; median:4.0, IQR:2.5–9 (months)) and 3767 (age at onset as phenotype; median:67, IQR:61–73 (years)). cases of probable or definite sCJD by contemporary diagnostic criteria either included in a previous paper from the collaborative group [19], or newly genotyped on Illumina’s Global Screening Array (Table 1). All patients were deceased.
Table 1. Number of samples used in the association test from 12 countries (duration / age) with interquartile range and median.
| Country | N (duration) | N (age) | Median (duration) | IQR (duration) | Median (age) | IQR (age) |
|---|---|---|---|---|---|---|
| Australia | 22 | 22 | 2.05 | 1.87 | 67 | 12.5 |
| Austria | 44 | 44 | 4.5 | 5.87 | 72 | 7 |
| Canada | 133 | 133 | 4 | 5 | 67 | 14 |
| France | 95 | 95 | 4 | 4 | 68 | 13 |
| Germany | 798 | 792 | 6 | 8 | 66 | 12 |
| Italy | 554 | 554 | 5 | 7 | 67 | 13 |
| Netherlands | 126 | 126 | 3.94 | 4.02 | 66.5 | 13 |
| Poland | 42 | 42 | 3 | 2.88 | 63.5 | 9.25 |
| Spain | 74 | 74 | 3.45 | 4.25 | 69 | 13.75 |
| Switzerland | 35 | 35 | 2.69 | 2.91 | 70 | 13.5 |
| UK | 951 | 951 | 5 | 6.95 | 67 | 12 |
| USA | 899 | 899 | 3 | 5 | 67 | 13 |
| Total | 3773 | 3767 |
Genotype doses were imputed using the Michigan Imputation Server [22], resulting in 6,308,901 SNPs passing quality control.
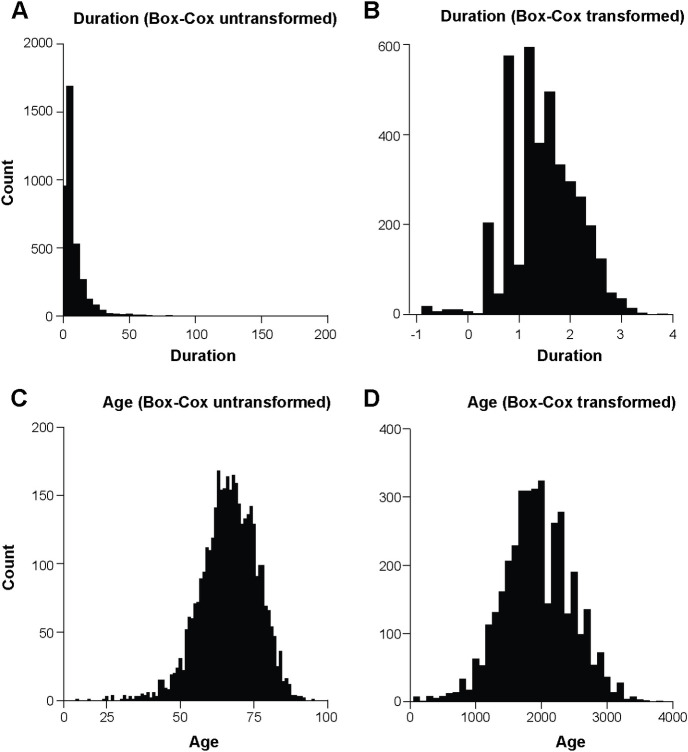
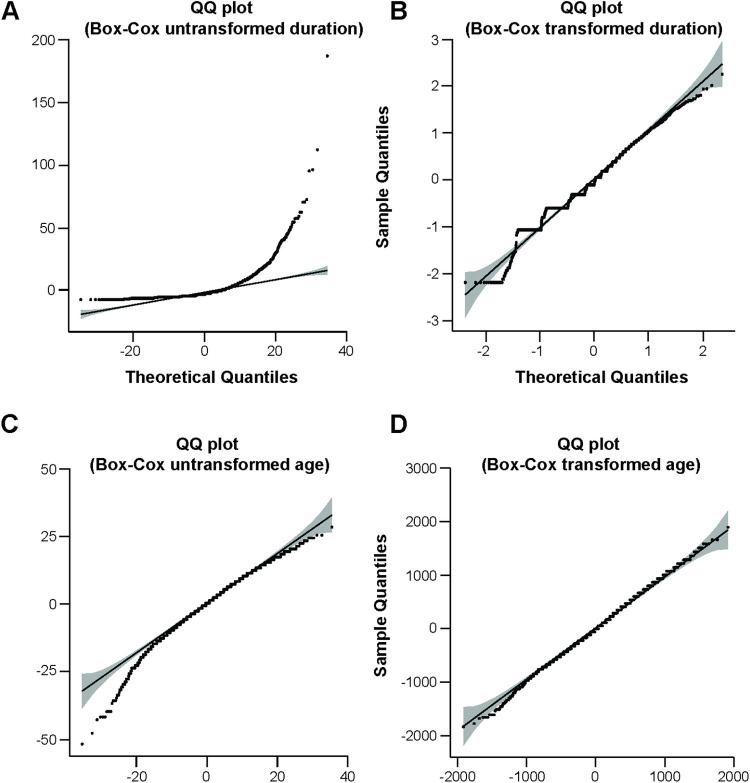
The median age / duration for men was 67 years and 3.8 months respectively and 67 years and 4.0 months for women. Median clinical duration (2.0–6.0 months) and age at onset (63.5–72 years) varied by site, so this was included as a covariate in the analysis. Phenotypes were modelled as normally distributed quantitative traits following transformation using methods developed by Box and Cox [26] illustrated as histograms and QQ plots (Figs 1 and 2; S1 Fig). Association analysis omitting sex, age or country or any combination as covariates did not show any significant difference in terms of outcome. Principal components analysis was used to exclude cases with distinct ancestry (n = 54) and did not suggest any strong effects of ancestry on the outcomes of interest (S2 and S3 Figs).
Fig 1.
Histograms for phenotypes duration before (A) and after (B) Box-Cox transformation and age before (C) and after (D) Box-Cox transformation.
Fig 2.
Quantile-Quantile plots for phenotypes duration before (A) and after (B) Box-Cox transformation and age before (C) and after (D) Box-Cox transformation.
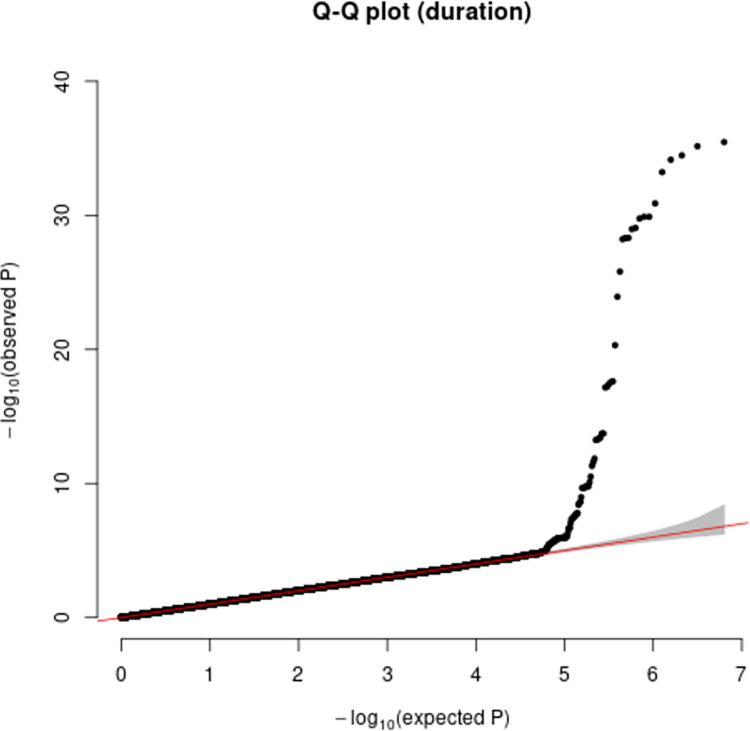
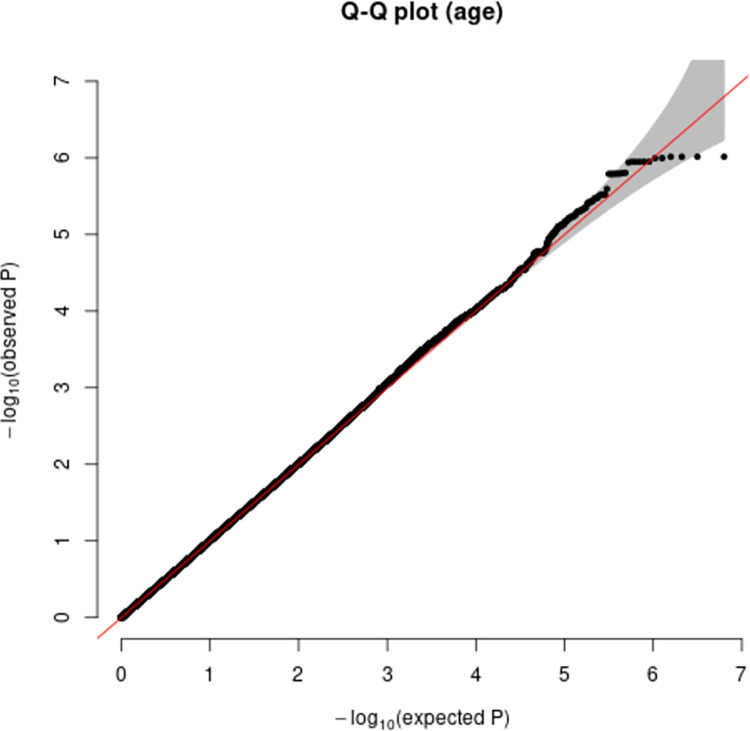
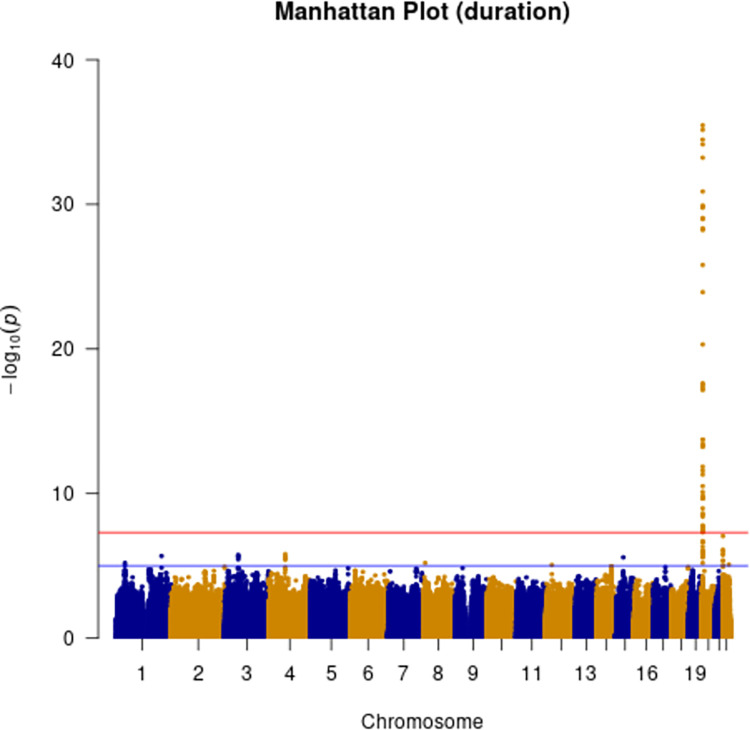
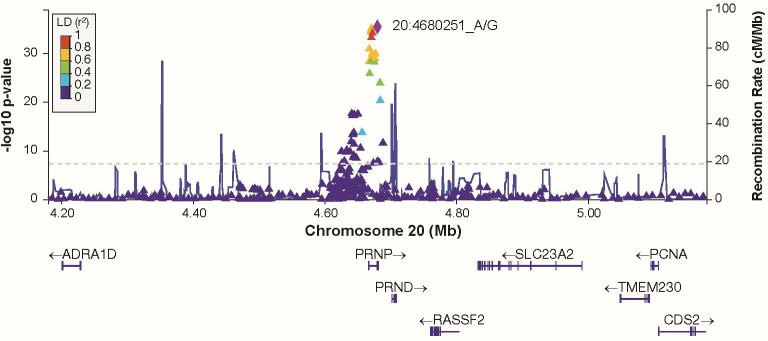
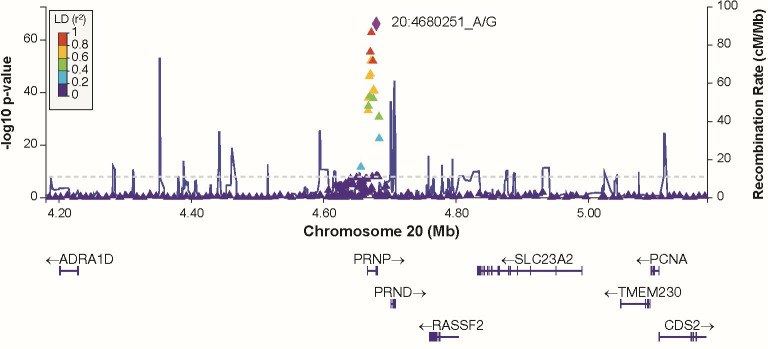
Additive and heterozygous genetic models were run genome-wide in SNPTEST with sex, contributing site and genetic ancestry covariates (see Methods) without any statistical inflation (lambda = 1.000 / 1.000 for clinical duration / age) as illustrated with QQ plots in Fig 3 (duration phenotype) and Fig 4 (age phenotype). 53 SNPs achieved genome-wide significance (P<5x10-8) for the clinical duration phenotype (additive model) (Fig 5 and S1 Table), all at the PRNP locus (top SNP rs1799990, pvalue = 3.45x10-36, beta = 0.34 for additive model; rs1799990, pvalue = 9.92x10-67, beta = 0.84 for heterozygous model, Figs 6 and 7). PRNP rs1799990 was the obvious outstanding genome-wide candidate determinant of clinical duration.
Fig 3. Quantile-Quantile plot with duration as phenotype.
Fig 4. Quantile-Quantile plot with age as phenotype.
Fig 5. Manhattan plot with clinical duration as phenotype.
(red line indicating genome-wide significance of 5x10-8; blue line indicating suggestive genome-wide significance (5x10-8 > pvalue < 1x10-5)).
Fig 6. Regional association plot at PRNP locus with clinical duration as phenotype (additive model).
Fig 7. Regional association plot at PRNP locus with clinical duration as phenotype (heterozygous model).
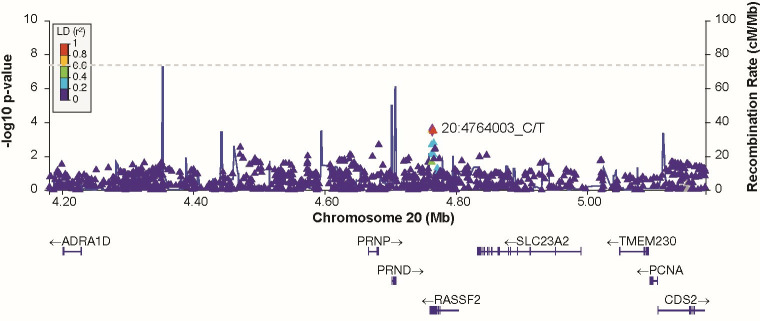
Of 68 cis-eQTL SNPs associated with PRNP expression in various brain tissues (obtained from GTEx); none were present in the list of 53 SNPs achieving genome-wide significance (duration phenotype). 50 of these eQTL SNPs for PRNP passed QC, all were P>0.001 (duration phenotype). No genome-wide significant SNPs remained after conditioning for rs1799990 codon 129 (Fig 8 and S4 Fig). There were 51 suggestive associated SNPs (5x10-8 > pvalue<1x10-5, including at regions near to HDHD5 (chromosome 22), FHIT (chromosome 3) and EREG (chromosome 4) (S2 Table and S5–S7 Figs). There were no significant gene-based associations of clinical duration apart from PRNP (MAGMA and FUMA) (Tables 2 and 3).
Fig 8. Regional association plot at PRNP locus for conditional analysis on SNP rs1799990 with clinical duration as phenotype (additive model).
Table 2. Top 10 genes identified by MAGMA (standalone) gene analysis (including genome-wide significant SNPs) with duration as phenotype.
| Gene | NCBI Gene ID | Chr | Start (hg19) | Stop (hg19) | NSNPS | N | ZSTAT | Pvalue | Bonf. corr. Pvalue |
|---|---|---|---|---|---|---|---|---|---|
| PRNP | 5621 | 20 | 4641797 | 4707235 | 189 | 3773 | 6.11 | 5.00 x 10−10 | 9.02 x 10−6 |
| ANP32E | 81611 | 1 | 150165717 | 150233504 | 82 | 3773 | 4.15 | 1.64 x 10−5 | 0.31 |
| CA14 | 23632 | 1 | 150204554 | 150262478 | 59 | 3773 | 3.92 | 4.38 x 10−5 | 0.79 |
| TMEM121B | 27439 | 22 | 17572189 | 17627257 | 148 | 3773 | 3.90 | 4.81 x 10−5 | 0.87 |
| HDHD5 | 27440 | 22 | 17593410 | 17671177 | 299 | 3773 | 3.89 | 5.10 x 10−5 | 0.92 |
| IL17RA | 23765 | 22 | 17540849 | 17621584 | 185 | 3773 | 3.85 | 5.98 x 10−5 | 1 |
| CNTN3 | 5067 | 3 | 74286719 | 74688587 | 845 | 3773 | 3.58 | 1.70 x 10−4 | 1 |
| APH1A | 51107 | 1 | 150212799 | 150266609 | 57 | 3773 | 3.54 | 2.03 x 10−4 | 1 |
| U2SURP | 23350 | 3 | 142695366 | 142804567 | 170 | 3773 | 3.50 | 2.36 x 10−4 | 1 |
| FZD8 | 8325 | 10 | 35902177 | 35955362 | 83 | 3773 | 3.47 | 2.58 x 10−4 | 1 |
(NSNPS = number of SNPs annotated to a gene; N = number of samples; ZSTAT = Z-score for the gene, based on its p-value)
Table 3. Top 10 genes identified by FUMA gene analysis (including genome-wide significant SNPs) with duration as phenotype.
| Gene | Chr | Start (hg19) | Stop (hg19) | NSNPS | N | ZSTAT | Pvalue | Bonf. corr. Pvalue |
|---|---|---|---|---|---|---|---|---|
| PRNP | 20 | 4666882 | 4682236 | 27 | 3773 | 7.02 | 1.12 x 10−12 | 2.03 x 10−8 |
| CECR5 | 22 | 17618401 | 17646177 | 81 | 3773 | 4.28 | 9.42 x 10−6 | 0.17 |
| ANP32E | 1 | 150190717 | 150208504 | 19 | 3773 | 3.86 | 5.63 x 10−5 | 1 |
| CA14 | 1 | 150229554 | 150237478 | 3 | 3773 | 3.79 | 7.60 x 10−5 | 1 |
| AL356356.1 | 1 | 150521897 | 150524367 | 1 | 3773 | 3.67 | 1.23 x 10−4 | 1 |
| AC006946.15 | 22 | 17602476 | 17612994 | 40 | 3773 | 3.51 | 2.20 x 10−4 | 1 |
| CCDC174 | 3 | 14693271 | 14714166 | 66 | 3773 | 3.51 | 2.26 x 10−4 | 1 |
| KCNJ3 | 2 | 155554811 | 155714863 | 453 | 3773 | 3.45 | 2.82 x 10−4 | 1 |
| VRK3 | 19 | 50479724 | 50529203 | 115 | 3773 | 3.38 | 3.56 x 10−4 | 1 |
| U2SURP | 3 | 142683339 | 142779567 | 154 | 3773 | 3.38 | 3.61 x 10−4 | 1 |
(NSNPS = number of SNPs annotated to a gene; N = number of samples; ZSTAT = Z-score for the gene, based on its p-value)
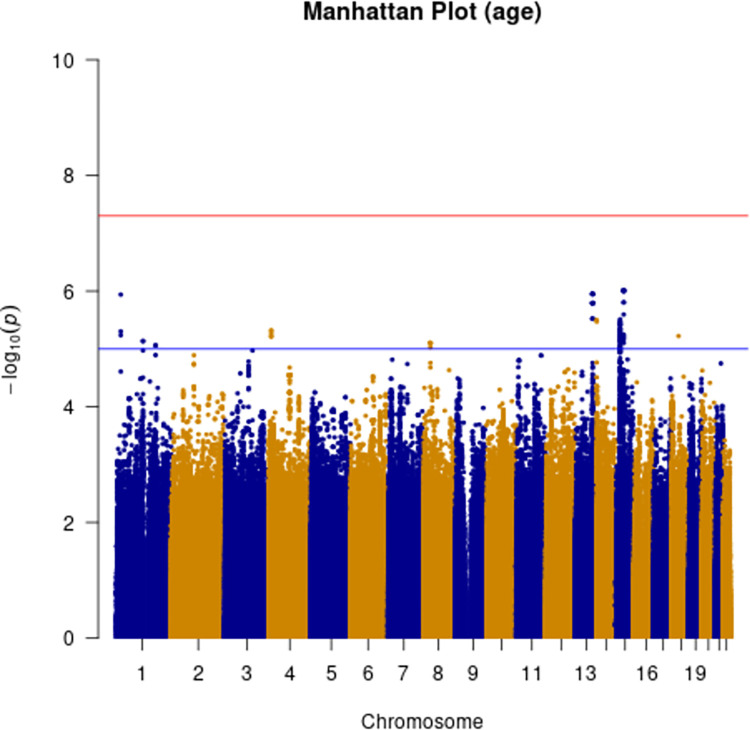
Age-based analysis did not identify any genome-wide significant SNP associations (Fig 9). Two suggestive associations were identified on chromosome 15 near NEDD4 and chromosome 13 near UGGT2 (S8 and S9 Figs). Gene-based analysis for age at onset with MAGMA identified HS6ST3 (pvalue = 1.93 x 10−6), with similarly significant association detected using FUMA (S3 and S4 Tables).
Fig 9. Manhattan plot with age at onset as phenotype.
(red line indicating genome-wide significance of 5x10-8; blue line indicating suggestive genome-wide significance (5x10-8 > pvalue < 1x10-).
Gene-set analysis for clinical duration using FUMA (including PRNP locus) identified binders of type-5 metabotropic glutamate receptors (GO Molecular Function ontology n = 1738, pvalue = 1.85 x 10−5) (Tables 4 and 5). Gene-set analysis for age at onset using MAGMA revealed intracellular oxygen homeostasis as a significant term (pvalue = 1.89 x 10−6) (S5 Table). Genetic correlation between clinical duration GWAS and the previously published case-control GWAS resulted in a non-significant genetic correlation of 0.1467 (pvalue = 0.79, 95% CI 0.92,1.21; S6 Table). Meta-analysis of the two GWAS (case-only and case-control) resulted in the same strong codon 129 effect as described above whilst removing the suggestive locus on chromosome 22 the HDHD5 locus (S10 Fig).
Table 4. Top 10 pathways identified by MAGMA (standalone) gene-set analysis (including genome-wide SNPs) using duration as phenotype.
| Category | Pathway | NGENES | BETA | Pvalue | Bonf. Corr. Pvalue |
|---|---|---|---|---|---|
| Gene ontology | regulation of calcium ion import across plasma membrane | 2 | 2.86 | 2.57 x 10−6 | 0.04 |
| Gene ontology | regulation of T-lymphocyte activation via T cell receptor contact with MHC-bound antigen | 5 | 1.58 | 5.41 x 10−6 | 0.09 |
| Gene ontology | cellular response to copper | 11 | 1.04 | 2.55 x 10−5 | 0.43 |
| Gene ontology | proteosomal ubiquitin-independent protein catabolic process | 4 | 1.51 | 6.25 x 10−5 | 1.00 |
| Gene ontology | anchored component of external side of plasma membrane | 18 | 0.77 | 1.06 x 10−4 | 1 |
| Gene ontology | response to iron ion | 30 | 0.61 | 1.11 x 10−4 | 1 |
| Gene ontology | obsolete intrinsic component of external side of plasma membrane | 23 | 0.69 | 1.16 x 10−4 | 1 |
| Gene ontology | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell | 8 | 1.06 | 1.27 x 10−4 | 1 |
| Gene ontology | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation | 4 | 1.33 | 3.44 x 10−4 | 1 |
| Gene ontology | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell | 2 | 1.67 | 3.52 x 10−4 | 1 |
(NGENES = number of genes in the gene-set dataset; BETA = regression coefficient of the gene set)
Table 5. Top 10 pathways identified by FUMA gene-set analysis (including genome-wide SNPs) using duration as phenotype.
| Category | Pathway | NGENES | BETA | Pvalue | Bonf. corr. Pvalue |
|---|---|---|---|---|---|
| Gene ontology | type_5_metabotropic_glutamate_receptor_binding | 5 | 1.92 | 1.85 x 10−5 | 0.29 |
| Gene ontology | ureteric_bud_elongation | 9 | 1.00 | 8.58 x 10−5 | 1 |
| Gene ontology | negative_regulation_of_cell_maturation | 8 | 0.89 | 1.53 x 10−5 | 1 |
| Gene ontology | mechanosensory_behavior | 13 | 0.85 | 1.55 x 10−5 | 1 |
| Gene ontology | actin_filament_based_transport | 8 | 0.90 | 3.84 x 10−5 | 1 |
| Gene ontology | learned_vocalization_behavior_or_vocal_learning | 8 | 0.97 | 3.99 x 10−5 | 1 |
| Gene ontology | peptidyltransferase_activity | 3 | 1.70 | 5.78 x 10−5 | 1 |
| Gene ontology | pyrimidine_containing_compound_transmembrane_transport | 10 | 0.79 | 7.38 x 10−5 | 1 |
| Curated gene sets | smid_breast_cancer_relapse_in_pleura_dn | 24 | 0.49 | 7.89 x 10−5 | 1 |
| Gene ontology | vitamin_binding | 128 | 0.23 | 8.75 x 10−4 | 1 |
(NGENES = number of genes in the gene-set dataset; BETA = regression coefficient of the gene set)
We also calculated the power of the study based on 3773 samples and a genome-wide significance level of 5x10-8 using the additive model with a range of effect sizes and minor allele frequencies. Plotting the most significant SNP (PRNP; rs1799990) and the lead SNPs of the suggestive association signals (HDHD5, rs4819962; FHIT, rs2366847; EREG, rs11727991) resulted in rs1799990 achieving full power and the three lead SNPs being borderline achieving a power value of ~0.7–0.8 (S11 Fig).
Interestingly, there was no evidence that the sCJD genetic susceptibility genes, STX6 or GAL3ST1, which were identified in the previously published case-control study [19], modify clinical phenotypes. The identification of these genes in the case-control GWAS implicated intracellular trafficking and sphingolipid metabolism respectively as causal disease mechanisms. To further investigate the roles of these pathways in disease phenotypes, we compiled a comprehensive, bespoke gene list including genes related to these pathways, which have been implicated in neurodegenerative diseases, and performed MAGMA analysis (S7 and S8 Tables). This highlighted UGGT2, a sphingolipid metabolism linked gene, to be associated with sCJD age of onset.
Discussion
We describe the first well-powered GWAS for phenotypic traits in sporadic human prion disease. The only clearly identified risk locus was the PRNP gene itself, more specifically the well-known common variant at codon 129, for the clinical duration phenotype. Conditioning for the codon 129 polymorphism at this locus removed all evidence of association at the locus, implicating the coding sequence of PRNP and not PrP expression in controlling this phenotype. We found a number of suggestive risk loci with P<10−5, which should require additional genetic evidence before being considered further. Pathway analysis identified binders of type-5 metabotropic glutamate receptors, which are known to mediate the downstream effects of amyloid beta bound to prion protein, as a top hit for clinical duration [27, 28]. Importantly however, since this small gene set (n = 5) was non-significant after removing PRNP, these data should be interpreted with caution. Overall, this work further establishes the key importance of the PrP coding sequence relative to other potential mechanisms and genetic loci in determination of CJD survival.
For age at onset there were no genome-wide significant SNPs, but we identified the HS6ST3 in a gene-based test and intracellular oxygen homeostasis by pathway analysis (S3–S5 Tables). HS6ST3 or Heparan Sulfate 6-O-Sulfotransferase 3 catalyses the transfer of sulfate from 3’-phosphoadenosine 5’-phosphosulfate (PAPS) to position 6 of the N-sulfoglucosamine residue (GlcNS) of heparan sulfate (HS), thus potentially modifying the interactions of this molecule with cell surface proteins. There is a vast literature on a role for polyanionic compounds, including HS in prion disease pathogenesis, as they colocalise with PrPC on the cell surface and with aggregated PrPSc [29], act as potential co-factors in prion replication, and there is potent inhibitory activity of HS and related compounds on prion propagation [30]. A role for intracellular oxygen homeostasis is less clearly linked to prion disease. Both associations were borderline in significance taking into account multiple testing. We found no evidence of genetic correlation between the case-only and published case-control GWAS analyses. We observed only a moderate heritability (h2SNP = 0.18–0·26, using different methods) for the case-control GWAS [19], and low heritability for the duration phenotype (h2SNP = 0.09 using LDSC). Common SNPs measured in these studies therefore explain only a small proportion of disease phenotypes. The only locus common to both GWAS studies is PRNP, with no evidence that SNPs at the STX6 or GAL3ST1 loci have any effect on clinical phenotypes in lead SNP association, gene-based or pathway analyses. It is possible that larger sample sizes, with additional risk factor discovery, will uncover shared determinants, but the current evidence suggests that beyond PRNP, distinct mechanisms and/or stochasticity determines disease risk, age at onset and clinical duration.
Absence of an association between PRNP cis-eQTL SNPs and clinical duration/age of onset should not deter the pursuit of methods to reduce PrP as a therapeutic strategy. There is a wealth of evidence for the safety and potential effectiveness of this approach from animal models [31–35]. PRNP cis-eQTL SNPs are predominantly associated with localised tissue expression of PrP, typically in cerebellum or cerebellar hemispheres, and are relatively modest effects. Therapeutic strategies aim for more profound protein knock-down, which will be critical to achieve across a wide range of central nervous system tissues and cell types [36].
Poleggi et al. (2018) [37] aimed to identify additional genetic modifiers in a GWAS study with a small cohort of patients (E200K mutation only). In this study, two SNPs were identified within the CYP4X1 gene locus indicating that this gene modulates onset of disease in sCJD. The top SNP identified in the Poleggi analysis (rs9793471) had a pvalue of 0.08 in our analysis.
A number of GWAS studies reporting genetic modifiers in other neurological diseases of in relation to the age at onset phenotype have been reported. One example is the case-only study of Li et al. [38] where a number of novel genes for age-at onset in Alzheimer’s disease were identified. Blauwendraat et al. [39] described several modifier loci in an age-at-onset GWAS analysis of Parkinson’s disease.
It was imperative to transform the non-normal distribution of the duration phenotype data as the GWAS association model requires Gaussian distributed phenotype data to avoid model misspecification, which could lead to false conclusions. A number of data transformations were tested (log, rank inverse, square root) for transformation of the phenotype data (duration and age) and the Box-Cox transformation was found to be the best option for establishing the optimal correlation coefficient ensuring a normal distribution and reduction of data noise to a minimum.
This study was limited by sample size and was restricted to the examination of age at onset and clinical duration phenotypes that are almost universally collected, whereas the diversity of clinical phenotypes in CJD is well known (including variable involvement of cognitive, ataxic, psychiatric, sleep and motor aspects). In biochemical aspects and biomarkers, we see diversity of PrPSc types, and different imaging, neurophysiological and fluid biomarker associations. These parameters are only collected in smaller subsets of data. Genetic studies in a rare disease like sCJD benefit from national investment and collaboration in prion disease surveillance [40]. Future work of the collaborative group might focus on building larger sample collections for increased power, exome or genome studies to ascertain rare and structural variants and extension of these type of analyses to other phenotypes (e.g., the well-known subtypes of CJD based on major symptom at presentation (ataxia, visual processing disorder etc.)).
Supporting information
(DOCX)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Green rectangles indicate genes involved in neurological diseases (PrD = Prion Disease, PD = Parkinson’s Disease, AD = Alzheimer’s Disease, ALS = Amyotrophic Lateral Sclerosis; HD = Huntington’s Disease, FTD = Frontotemporal Dementia) (NSNPS = number of SNPs annotated to a gene; N = number of samples; ZSTAT = Z-score for the gene, based on its p-value).
(XLSX)
Green rectangles indicate genes involved in neurological diseases (PrD = Prion Disease, PD = Parkinson’s Disease, AD = Alzheimer’s Disease, ALS = Amyotrophic Lateral Sclerosis; HD = Huntington’s Disease, FTD = Frontotemporal Dementia). (NSNPS = number of SNPs annotated to a gene; N = number of samples; ZSTAT = Z-score for the gene, based on its p-value).
(XLSX)
(XLSX)
Acknowledgments
We thank Richard Newton for support with images and UCL Genomics who did the array processing. For UK samples we would like to thank patients, their families and carers, UK neurologists and other referring physicians, co-workers at the National Prion Clinic, our colleagues at the National Creutzfeldt-Jakob Disease Research and Surveillance Unit, Edinburgh. We thank Dr. Maria Styczynska from Mossakowski Medical Research Centre; Polish Academy of Sciences; Warsaw, for kindly providing control DNA samples for the Polish cohort. We thank Inés Santiuste and the Valdecilla Biobank (PT17/0015/0019), integrated in the Spanish Biobank Network, for their support and collaboration in sample collection and management. We thank Megan Casey for assistance with sample collection and management. The Australian National Creutzfeldt-Jakob Disease Registry (ANCJDR) would like to thank all patients and their families for supporting surveillance activities that have allowed participation in the study, as well as their managing physicians. The Italian Creutzfeldt-Jakob Registry would like to thank Dr.Clara Salciccia for the collaboration in DNA sample collection and management, Cinzia Gasparrini for administrative assistance and all patients, their families, neurologists, and referring physicians.
Data Availability
The data used in this study have been deposited in the public database GWAS Catalogue (https://www.ebi.ac.uk/gwas/; accession number GCP000963).
Funding Statement
This work was supported by the MRC (UK) core grant to the MRC Prion Unit at UCL (code MC_UU_00024/1). Several authors at UCL/UCLH receive funding from the Department of Health’s NIHR Biomedical Research Centres funding scheme. Some of this work was supported by the Department of Health funded National Prion Monitoring Cohort study. Funding for the collection of Polish samples for study was partially provided by the EU joint programme JPND and Medical University of Lodz. The Italian national surveillance of Creutzfeldt-Jakob disease and related disorders is partially supported by the Ministero della Salute, Italy. The German National Reference Centre for TSE is funded by grants from the Robert-Koch-Institute. The Dutch National Prion Disease Registry is funded by the National Institute for Public Health and the Environment (RIVM), which is part from the Ministry for Health, Welfare and Sports, The Netherlands. PS-J was supported by Instituto de Salud Carlos III [Fondo de Investigación Sanitaria, PI16/01652] Accion Estrategica en Salud integrated in the Spanish National I+D+i Plan and financed by Instituto de Salud Carlos III (ISCIII) – Subdireccion General de Evaluacion and the Fondo Europeo de Desarrollo Regional (FEDER – “Una Manera de Hacer Europa”). The French National Surveillance Network for Creutzfeldt-Jakob disease is supported by Santé Publique France. MDG (UCSF) receives research support from the NIH/NIA (grant R01 AG031189, R56AG055619, R01AG062562) and the Michael J. Homer Family Fund. The National Prion Disease Pathology Surveillance Center in the U.S. is funded by the Centers for Disease Control and Prevention (NU38CK000486). The Austrian Reference Center for Human Prion diseases (ÖRPE) is supported by the Austrian Ministy of Health – Bundesministerium für Soziales, Gesundheit, Pflege und Konsumentenschutz. The ANCJDR is supported through funding from the Commonwealth Department of Health and Aged Care.
References
- 1.Klug GM, Wand H, Simpson M, Boyd A, Law M, Masters CL, et al. Intensity of human prion disease surveillance predicts observed disease incidence. J Neurol Neurosurg Psychiatry 2013; 84(12): 1372–1377. doi: 10.1136/jnnp-2012-304820 [DOI] [PubMed] [Google Scholar]
- 2.Pocchiari M, Puopolo M, Croes EA, Budka H, Gelpi E, Collins S, et al. Predictors of survival in sporadic Creutzfeldt-Jakob disease and other human transmissible spongiform encephalopathies. Brain. 2004; 127: 2348–2359. doi: 10.1093/brain/awh249 [DOI] [PubMed] [Google Scholar]
- 3.Thompson AG, Lowe J, Fox Z, Lukic A, Porter MC, Ford L, et al. The Medical Research Council prion disease rating scale: a new outcome measure for prion disease therapeutic trials developed and validated using systematic observational studies. Brain. 2013; 136(Pt 4): 1116–1127. doi: 10.1093/brain/awt048 [DOI] [PubMed] [Google Scholar]
- 4.Nihat A, Mok TH, Odd H, Thompson AGB, Caine D, McNiven K, et al. Development of novel clinical examination scales for the measurement of disease severity in Creutzfeldt-Jakob disease. J Neurol Neurosurg Psychiatry. 2022; 93(4): 404–412. doi: 10.1136/jnnp-2021-327722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tam J, Centola J, Kurudzhu H, Watson N, MacKenzie J, Leitch M, et al. Sporadic Creutzfeldt-Jakob Disease in the young (50 and below): 10-year review of United Kingdom surveillance. J Neurol. 2023; 270(2): 1036–1046. doi: 10.1007/s00415-022-11467-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Appleby BS, Maddox R, Schonberger LB, Cali I, Hammett T, Cohen M, et al. Sporadic Creutzfeldt-Jakob Disease in a Very Young Person. Neurology. 2021; 97(17): 813–816. doi: 10.1212/WNL.0000000000012737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.NCJDRSU (The National CJD Research & Surveillance Unit). Annual Report 2021. [cited 2022. Available from: https://www.cjd.ed.ac.uk/sites/default/files/report29.pdf. [Google Scholar]
- 8.Nihat A, Ranson JM, Harris D, Mcniven K, Mok T, Rudge P, et al. Development of Prognostic Models for Survival and Care Status in Sporadic Creutzfeldt-Jakob disease. Brain Commun. 2022; 4(4); fcac201. doi: 10.1093/braincomms/fcac201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science. 1982; 216: 136–144. doi: 10.1126/science.6801762 [DOI] [PubMed] [Google Scholar]
- 10.Kraus A, Hoyt F, Schwartz CL, Hansen B, Artikis E, Hughson AG, et al. High-resolution structure and strain comparison of infectious mammalian prions. Mol Cell. 2021; 81(21): 4540–4551 e6. doi: 10.1016/j.molcel.2021.08.011 [DOI] [PubMed] [Google Scholar]
- 11.Manka SW, Zhang W, Wenborn A, Betts J, Joiner S, Saibil HR, et al. 2.7 A cryo-EM structure of ex vivo RML prion fibrils. Nat Commun. 2022; 13(1): 4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riek R, Hornemann S, Wider G, Billeter M, Glockshuber R, Wuthrich K. NMR structure of the mouse prion protein domain PrP(121–231). Nature. 1996; 382(6587): 180–182. doi: 10.1038/382180a0 [DOI] [PubMed] [Google Scholar]
- 13.Wadsworth JD, Asante EA, Collinge J. Contribution of transgenic models to understanding human prion disease. Neuropathol Appl Neurobiol. 2010; 36(7): 576–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manka SW, Wenborn A, Betts J, Joiner S, Saibil HR, Collinge J, et al. A structural basis for prion strain diversity. Nat Chem Biol. 2023; 19(5): 607–613. doi: 10.1038/s41589-022-01229-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mercer RCC, Harris DA. Mechanisms of prion-induced toxicity. Cell Tissue Res. 2023; 392(1): 81–96. doi: 10.1007/s00441-022-03683-0 [DOI] [PubMed] [Google Scholar]
- 16.Lakkaraju AKK, Frontzek K, Lemes E, Herrmann U, Losa M, Marpakwar R, et al. Loss of PIKfyve drives the spongiform degeneration in prion diseases. EMBO Mol Med. 2021; 13(9): e14714. doi: 10.15252/emmm.202114714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moreno JA, Radford H, Peretti D, Steinert JR, Verity N, Martin MG, et al. Sustained translational repression by eIF2 alpha-P mediates prion neurodegeneration. Nature. 2012; 485(7399): 507–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Llorens F, Rubsamen N, Hermann P, Schmitz M, Villar-Pique A, Goebel S, et al. A prognostic model for overall survival in sporadic Creutzfeldt-Jakob disease. Alzheimers Dement. 2020; 16(10): 1438–1447. doi: 10.1002/alz.12133 [DOI] [PubMed] [Google Scholar]
- 19.Jones E, Hummerich H, Vire E, Uphill J, Dimitriadis A, Speedy H, et al. Identification of novel risk loci and causal insights for sporadic Creutzfeldt-Jakob disease: a genome-wide association study. Lancet Neurol. 2020; 19(10): 840–848. doi: 10.1016/S1474-4422(20)30273-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hermann P, Appleby B, Brandel JP, Caughey B, Collins S, Geschwind MD, et al. Biomarkers and diagnostic guidelines for sporadic Creutzfeldt-Jakob disease. Lancet Neurol. 2021; 20(3): 235–246. doi: 10.1016/S1474-4422(20)30477-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mastrangelo A, Mammana A, Baiardi S, Tiple D, Colaizzo E, Rossi M, et al. Evaluation of the impact of CSF prion RT-QuIC and amended criteria on the clinical diagnosis of Creutzfeldt-Jakob disease: a 10-year study in Italy. J Neurol Neurosurg Psychiatry. 2023; 94(2): 121–129. doi: 10.1136/jnnp-2022-330153 [DOI] [PubMed] [Google Scholar]
- 22.Das S, Forer L, Schonherr S, Sidore C, Locke AE, Kwong A, et al. Next-generation genotype imputation service and methods. Nat Genet. 2016; 48(10): 1284–1287. doi: 10.1038/ng.3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bulik-Sullivan B, Loh P, Finucane H, Ripke S, Yang J, Schizophrenia Working Group of the Psychiatric Genomics Consortium et al. LD Score Regression Distinguishes Confounding from Polygenicity in Genome-Wide Association Studies. Nat Genet. 2015; 47(3): 291–295. doi: 10.1038/ng.3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017; 8(1): 1826. doi: 10.1038/s41467-017-01261-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015; 11(4): e1004219. doi: 10.1371/journal.pcbi.1004219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Box GEP, Cox DR. An analysis of transformations. Journal of Royal Statistical Society; Series B. 1964; 26: 211–243. [Google Scholar]
- 27.Um JW, Kaufman AC, Kostylev M, Heiss JK, Stagi M, Takahashi H, et al. Metabotropic glutamate receptor 5 is a coreceptor for Alzheimer abeta oligomer bound to cellular prion protein. Neuron. 2013; 79(5): 887–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haas LT, Salazar SV, Kostylev MA, Um JW, Kaufman AC, Strittmatter SM. Metabotropic glutamate receptor 5 couples cellular prion protein to intracellular signalling in Alzheimer’s disease. Brain. 2015; 139(2): 526–546. doi: 10.1093/brain/awv356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rouvinski A, Karniely S, Kounin M, Moussa S, Goldberg MD, Warburg G, et al. Live imaging of prions reveals nascent PrPSc in cell-surface, raft-associated amyloid strings and webs. J Cell Biol. 2014; 204(3): 423–441. doi: 10.1083/jcb.201308028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larramendy-Gozalo C, Barret A, Daudigeos E, Mathieu E, Antonangeli L, Riffet C, et al. Comparison of CR36, a new heparan mimetic, and pentosan polysulfate in the treatment of prion diseases. J Gen Virol. 2007; 88(Pt 3): 1062–1067. doi: 10.1099/vir.0.82286-0 [DOI] [PubMed] [Google Scholar]
- 31.Bueler H, Aguzzi A, Sailer A, Greiner RA, Autenried P, Aguet M, et al. Mice devoid of PrP are resistant to scrapie. Cell. 1993; 73(7): 1339–1347. doi: 10.1016/0092-8674(93)90360-3 [DOI] [PubMed] [Google Scholar]
- 32.Mallucci G, Dickinson A, Linehan J, Klohn PC, Brandner S, Collinge J. Depleting neuronal PrP in prion infection prevents disease and reverses spongiosis. Science. 2003; 302(5646): 871–874. doi: 10.1126/science.1090187 [DOI] [PubMed] [Google Scholar]
- 33.White MD, Farmer M, Mirabile I, Brandner S, Collinge J, Mallucci GR. Single treatment with RNAi against prion protein rescues early neuronal dysfunction and prolongs survival in mice with prion disease. Proc Natl Acad Sci USA. 2008; 105(29): 10238–10243. doi: 10.1073/pnas.0802759105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raymond GJ, Zhao HT, Race B, Raymond LD, Williams K, Swayze EE, et al. Antisense oligonucleotides extend survival of prion-infected mice. JCI Insight. 2019;5(16): e131175. doi: 10.1172/jci.insight.131175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minikel EV, Zhao HT, Le J, O’Moore J, Pitstick R, Graffam S, et al. Prion protein lowering is a disease-modifying therapy across prion disease stages, strains and endpoints. Nucleic Acids Res. 2020; 48(19): 10615–10631. doi: 10.1093/nar/gkaa616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mortberg MA, Gentile JE, Nadaf NM, Vanderburg C, Simmons S, Dubinsky D, et al. A single-cell map of antisense oligonucleotide activity in the brain. Nucleic Acids Res. 2023; 51(14): 7109–7124. doi: 10.1093/nar/gkad371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poleggi A, van der Lee S, Capellari S, Puopolo M, Ladogana A, De Pascali E, et al. Age at onset of genetic (E200K) and sporadic Creutzfeldt-Jakob diseases is modulated by the CYP4X1 gene. J Neurol Neurosurg Psychiatry. 2018;89(12):1243–1249 doi: 10.1136/jnnp-2018-318756 [DOI] [PubMed] [Google Scholar]
- 38.Li Y., Nuytemans K, La J, Jiang R, Slifer SH, Sun S et al. Identification of novel genes for age-at-onset of Alzheimer’s disease by combining quantitative and survival trait analyses. Alzheimers Dement 2023,;19(7): 3148–3157. doi: 10.1002/alz.12927 [DOI] [PubMed] [Google Scholar]
- 39.Blauwendraat C, Heilbron K, Vallerga CL, Bandres-Ciga S, von Coelln R, Pihlstrøm L et al. Parkinson’s disease age at onset genome-wide association study: Defining heritability, genetic loci, and α-synuclein mechanisms. Mov Disord. 2019; 34(6):866–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watson N, Brandel JP, Green A, Hermann P, Ladogana A, Lindsay T, et al. The importance of ongoing international surveillance for Creutzfeldt-Jakob disease. Nat Rev Neurol. 2021; 17(6): 362–379. doi: 10.1038/s41582-021-00488-7 [DOI] [PMC free article] [PubMed] [Google Scholar]