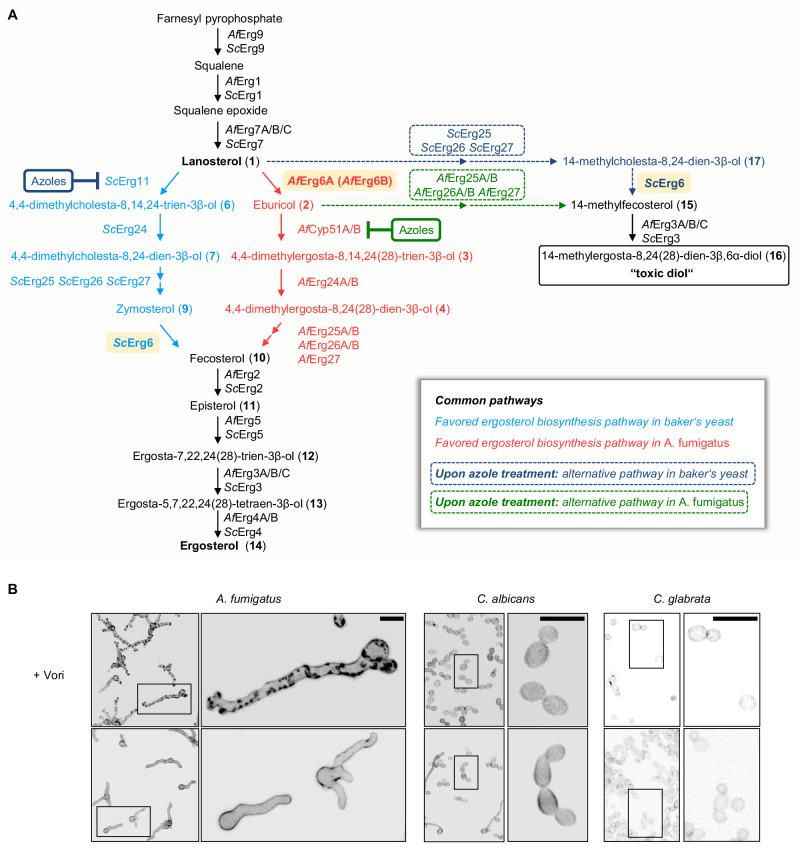
Fig. 1. The sterol biosynthesis pathway and the azole-induced formation of cell wall carbohydrate patches depends on the species.
A Ergosterol biosynthesis pathways in the mold A. fumigatus (Af; right pathway in red) and yeast S. cerevisiae (Sc; left pathway in light blue). In A. fumigatus, C24-methylation of lanosterol (1) by sterol C24-methyltransferase (AfErg6A; highlighted in light orange) is favored over C14-demethylation. In S. cerevisiae, lanosterol (1) is the favored substrate of sterol C14-demethylase (ScErg11). Both pathways converge with the formation of fecosterol (10), which is then further processed into ergosterol (14). Upon inhibition of sterol C14-demethylase (AfCyp51A/B and ScErg11) by azoles, alternative pathways may lead to the formation of a “toxic diol” (dashed arrows; alternative baker’s yeast pathway in dark blue, alternative A. fumigatus pathway in green). In S. cerevisiae, lanosterol (1) is converted to 14-methylcholesta-8,24-dien-3β-ol (17), which in turn is converted to 14-methylfecosterol (14-methylergosta-8,24(28)-dien-3β-ol) (15). In A. fumigatus, eburicol (2) is directly converted to 14-methylfecosterol (15). 14-methylfecosterol (15) is then converted to the 14-methylergosta-8,24(28)-dien-3β,6α-diol (16) which is considered a “toxic diol”. Ergosterol biosynthesis enzymes in A. fumigatus (Af) and S. cerevisiae (Sc): squalene synthase (AfErg9, ScErg9), squalene epoxidase (AfErg1, ScErg1), lanosterol synthase (AfErg7A/B/C, ScErg7), sterol C24-methyltransferase (AfErg6A, AfErg6B and ScErg6; highlighted in light orange), sterol C14-demethylase (AfCyp51A/B, ScErg11), sterol C14-reductase (AfErg24A/B, ScErg24), sterol C4-demethylase complex (AfErg25A/B, AfErg26A/B, AfErg27, ScErg25, ScErg26, ScErg27), sterol C8-isomerase (AfErg2, ScErg2), sterol C22-desturase (AfErg5, ScErg5), sterol C5-desaturase (AfErg3A/B/C, ScErg3), sterol C24 reductase (AfErg4A/B, ScErg4). B Conidia of A. fumigatus wild type and two Candida species (C. albicans ATCC14053 and C. glabrata ATCC2950) were inoculated in Sabouraud medium and incubated at 37 °C. After 9.5 h of incubation, the samples were either fixed and stored at 4 °C (control) or, after the medium was supplemented with 3 µg ml−1 voriconazole (+Vori), further incubated at 37 °C. After 15 h of additional incubation, the voriconazole-exposed hyphae and yeasts were also fixed. Samples were then stained with calcofluor white and analyzed with a confocal laser scanning microscope. Depicted are representative images of z-stack projections of optical stacks of the calcofluor white fluorescence covering the entire hyphae in focus. Upper panel, voriconazole-treated hyphae; lower panel, controls. Bars represent 10 μm. Data were representative of three experiments.